---
title: "Prof D's Regression Sessions - Vol 3"
subtitle: "AKA - In the (Linear) Mix(ed effects model practical for the next few hours)"
format:
html:
code-fold: true
code-tools: true
ipynb: default
filters:
- qna
- multicode
- quarto # keep Quarto’s built-ins as the last filter or it won't work
editor:
markdown:
wrap: 72
---
```{r}
#| message: false
#| warning: false
#| include: false
library(here)
here()
```
```{r}
#| message: false
#| warning: false
#| include: false
## Notes - make sure that reticulate is pointing to a local reticulate install of python or things might go squiffy.
## in terminal type: where python - to find out where reticulate might have stashed a version of the python.exe
## make sure you point to it before installing these packages using:
## reticulate::use_python("C:/Path/To/Your/Python/python.exe", required = TRUE)
#renv::install("C:/GitHubRepos/casaviz.zip")
# Get all packages from the lockfile
#lockfile <- renv::load("renv.lock")
#packages <- setdiff(names(lockfile$Packages), "casaviz")
# Restore only the selected packages
#renv::restore(packages = packages)
#Sys.unsetenv("VIRTUAL_ENV")
#Sys.setenv(RETICULATE_PYTHON = "C:/Users/adam_/anaconda3/envs/qmEnv/python.exe")
#library(reticulate)
# Point reticulate to the virtual environment's Python
#Sys.setenv(RETICULATE_PYTHON = "C:/GitHubRepos/QM_Fork/.venv/Scripts/python.exe")
# Check configuration
#py_config()
#library(reticulate)
#py_config()
library(reticulate)
#use_python("/QM_Fork/venv/Scripts/python.exe", required = TRUE)
#use_python("/GitHubRepos/QM_Fork/venv/Scripts/python.exe", required = TRUE)
# point reticulate to the right python installation - ideally the one reticulate installed.
#reticulate::use_python("C:/Users/Adam/AppData/Local/R/cache/R/reticulate/uv/cache/archive-v0/EiTNi4omakhlev5ckz2WP/Scripts/python.exe", required = TRUE)
#use_condaenv("qmEnv", conda = "C:/Users/adam_/anaconda3/Scripts/conda.exe", required = TRUE)
#use_condaenv("qmEnv", conda = "C:/Users/adam_/anaconda3/Scripts/conda.exe", required = TRUE)
#reticulate::use_python("C:/Users/adam_/anaconda3/envs/qmEnv/python.exe", required = TRUE)
#py_run_string("import pyproj; print(pyproj.CRS.from_epsg(27700))")
#virtualenv_create("r-reticulate", python = "C:/Users/Adam/AppData/Local/R/cache/R/reticulate/uv/cache/archive-v0/EiTNi4omakhlev5ckz2WP/Scripts/python.exe")
#virtualenv_install("r-reticulate", packages = c("pyyaml", "jupyter", "statsmodels","pyjanitor","pathlib","matplotlib","pandas", "numpy", "scipy", "seaborn", "geopandas", "folium", "branca"))
#use_virtualenv("r-reticulate", required = TRUE)
#py_install(packages = c(
# "pyyaml", "jupyter", "statsmodels", "pyjanitor", "pathlib",
# "matplotlib", "pandas", "numpy", "scipy", "seaborn",
# "geopandas", "folium", "branca", "plotly", "contexily"
#))
# reticulate::py_config()
# reticulate::py_require("pyyaml")
# reticulate::py_require("jupyter")
# reticulate::py_require("statsmodels")
# reticulate::py_require("pandas")
# reticulate::py_require("numpy")
# reticulate::py_require("pyjanitor")
# reticulate::py_require("pathlib")
# reticulate::py_require("matplotlib")
# reticulate::py_require("seaborn")
#reticulate::py_install("folium")
#reticulate::py_install("geopandas")
#reticulate::py_install("contextily", pip = TRUE)
#reticulate::py_install("scikit-learn", pip = TRUE)
```
```{=html}
<iframe data-testid="embed-iframe" style="border-radius:12px" src="https://open.spotify.com/embed/track/1mzn6ZJiEWwCsHTrr4ms0j?utm_source=generator" width="100%" height="352" frameBorder="0" allowfullscreen="" allow="autoplay; clipboard-write; encrypted-media; fullscreen; picture-in-picture" loading="lazy"></iframe>
```
## Preamble
We're going back in - Volume 3 of the Regression Sessions, so clearly we
are going to have to have volume 4 of the Progression Sessions. Yes,
"where is vol. 3?", I hear you ask. I don't know. Someone has lost it
and it's not up on Spotify, so we'll have to make do and jump to volume
4 - Enjoy!
## Practical Aims
1. Experiment with running some Linear Mixed Effects Models and
converting last week's show-stopper multiple regression model into a
Linear Mixed Effects version
2. Practice moving from an initial null model to a fully-fledged model
with fixed effects and random effects
3. Practice interpreting your final model and seeing if you can use it
to explain which local authority-level policy might be most
effective at raising attainment
## Read in your data
You can use your data from last week, but I've created a nice clean
dataset and added a few extra spicy variables about teachers and
schools' workforce in there for fun. All of that data is from here:
<https://explore-education-statistics.service.gov.uk/data-catalogue?themeId=b601b9ea-b1c7-4970-b354-d1f695c446f1&sortBy=newest&geographicLevel=SCH>
To save time, you might just want to download it directly from here:
https://github.com/adamdennett/QM/blob/main/sessions/L6_data/Performancetables_130242/2022-2023/school_data_2223.csv
::: multicode
#### {width="30"}
```{python}
import pandas as pd
import numpy as np
import janitor
from pathlib import Path
import statsmodels.api as sm
# little function to define the file root on different machines
def find_qm_root(start_path: Path = Path.cwd(), anchor: str = "QM") -> Path:
"""
Traverse up from the start_path until the anchor folder (e.g. 'QM' or 'QM_Fork') is found. Returns the path to the anchor folder.
"""
for parent in [start_path] + list(start_path.parents):
if parent.name == anchor:
return parent
raise FileNotFoundError(f"Anchor folder '{anchor}' not found in path hierarchy.")
qm_root = find_qm_root()
base_path = qm_root / "sessions" / "L6_data" / "Performancetables_130242" / "2022-2023"
na_all = ["", "NA", "SUPP", "NP", "NE", "SP", "SN", "LOWCOV", "NEW", "SUPPMAT", "NaN"]
qm_root = find_qm_root()
base_path = qm_root / "sessions" / "L6_data" / "Performancetables_130242" / "2022-2023"
na_all = ["", "NA", "SUPP", "NP", "NE", "SP", "SN", "LOWCOV", "NEW", "SUPPMAT", "NaN"]
england_filtered = pd.read_csv(base_path / "school_data_2223.csv", na_values=na_all, dtype={"URN": str})
# Log-transform safely: replace non-positive values with NaN
england_filtered['log_ATT8SCR'] = np.where(england_filtered['ATT8SCR'] > 0, np.log(england_filtered['ATT8SCR']), np.nan)
england_filtered['log_PTFSM6CLA1A'] = np.where(england_filtered['PTFSM6CLA1A'] > 0, np.log(england_filtered['PTFSM6CLA1A']), np.nan)
# Drop rows with NaNs in either column
england_filtered_clean = england_filtered.dropna(subset=['log_ATT8SCR', 'log_PTFSM6CLA1A'])
```
#### {width="41" height="30"}
```{r}
#| message: false
#| warning: false
library(tidyverse)
## Note edit your base path to refer to the folder where you have stored your data
base_path <- here("sessions", "L6_data", "Performancetables_130242", "2022-2023")
na_all <- c("", "NA", "SUPP", "NP", "NE", "SP", "SN", "LOWCOV", "NEW", "SUPPMAT")
england_filtered <- read_csv(file.path(base_path, "school_data_2223.csv"), na = na_all) |> mutate(URN = as.character(URN))
#str(england_filtered)
england_filtered_clean <- england_filtered[
!is.na(england_filtered$ATT8SCR) &
!is.na(england_filtered$PTFSM6CLA1A) &
england_filtered$ATT8SCR > 0 &
england_filtered$PTFSM6CLA1A > 0,
]
```
:::
## The Warm-up - Last week's Multiple Regression Show Stopper
- To get us going we are going to run our 'show-stopper' model from
last week so that we can compare this with a final linear mixed
effects version at the end of the practical.
- You should run your best model (without any interaction effects)
from last week, however, to help you along my best model from last
week looked like this:
::: multicode
#### {width="30"}
```{python}
import numpy as np
import statsmodels.formula.api as smf
# Filter rows where all log-transformed variables are > 0
england_filtered_clean = england_filtered_clean[
(england_filtered_clean['PTFSM6CLA1A'] > 0) &
(england_filtered_clean['PERCTOT'] > 0) &
(england_filtered_clean['PNUMEAL'] > 0)
].copy()
# Create log-transformed columns
england_filtered_clean['log_ATT8SCR'] = np.log(england_filtered_clean['ATT8SCR'])
england_filtered_clean['log_PTFSM6CLA1A'] = np.log(england_filtered_clean['PTFSM6CLA1A'])
england_filtered_clean['log_PERCTOT'] = np.log(england_filtered_clean['PERCTOT'])
england_filtered_clean['log_PNUMEAL'] = np.log(england_filtered_clean['PNUMEAL'])
# Fit the model
model = smf.ols(
formula='log_ATT8SCR ~ log_PTFSM6CLA1A + log_PERCTOT + log_PNUMEAL + OFSTEDRATING + gor_name + PTPRIORLO + ADMPOL_PT',
data=england_filtered_clean
).fit()
print(model.summary())
```
#### {width="41" height="30"}
```{r}
#| message: false
#| warning: false
england_filtered_clean <- england_filtered_clean %>%
filter(PTFSM6CLA1A > 0, PERCTOT > 0, PNUMEAL > 0)
england_model7 <- lm(log(ATT8SCR) ~ log(PTFSM6CLA1A) + log(PERCTOT) + log(PNUMEAL) + OFSTEDRATING + gor_name + PTPRIORLO + ADMPOL_PT, data = england_filtered_clean, na.action = na.exclude)
summary(england_model7)
```
:::
## Building Our Linear Mixed Effects Model
### Stage 1 - The Null / Variance Components Model for Ofsted Ratings
- If you remember from the lecture, in order to determine whether
running a linear mixed effects / multilevel model is necessary, if
you have categorical / grouping factors in your data set (**random
effects**), you should run the null model to see how much variance
the groups in your data are explaining.
- In our data, we had a couple of different grouping variable
candidates:
- Ofsted Rating (how 'good' or otherwise a school is as determined
by the Office for Standards in Education - Ofsted)
- Geographical groups. In our data these are Regions and the Local
Authorities within those regions.
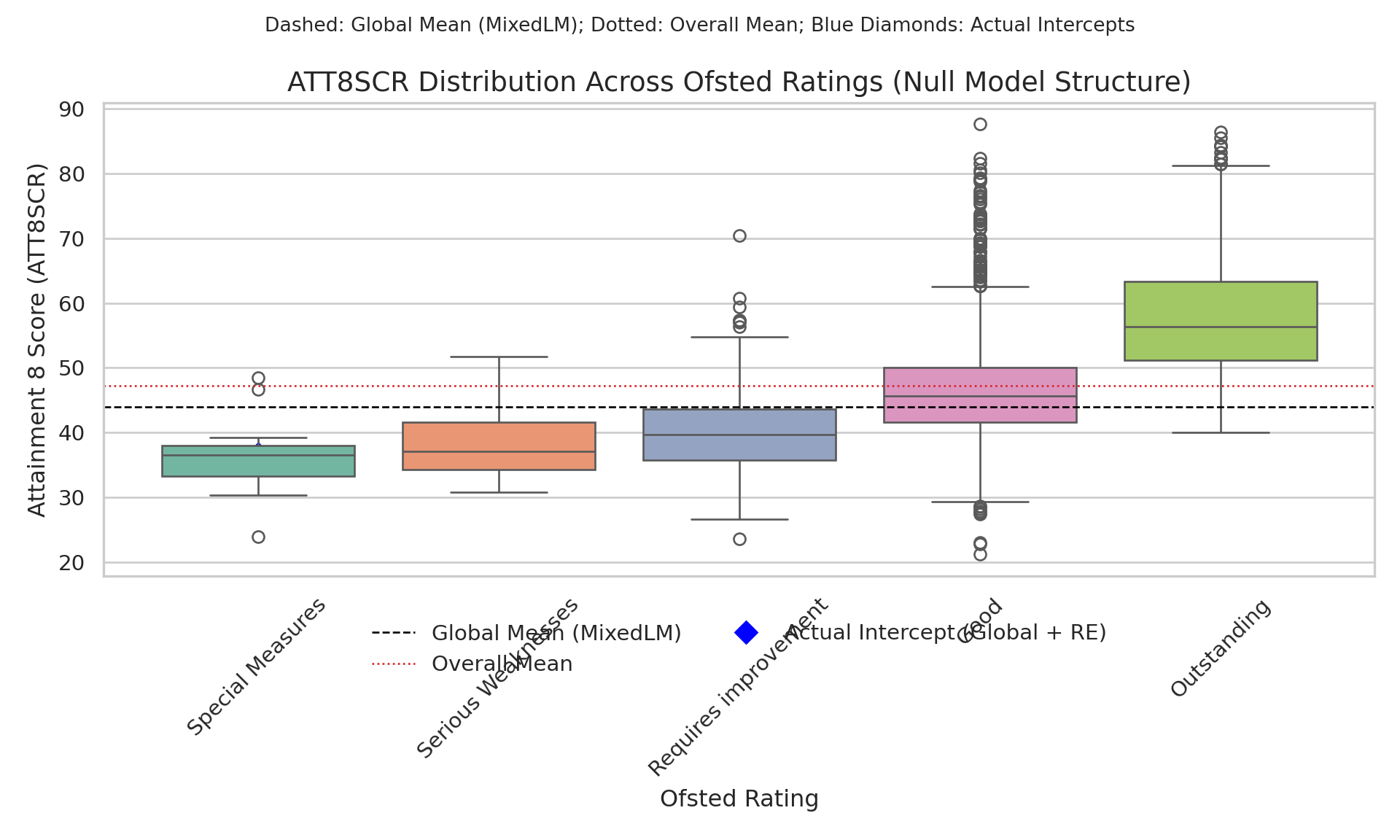
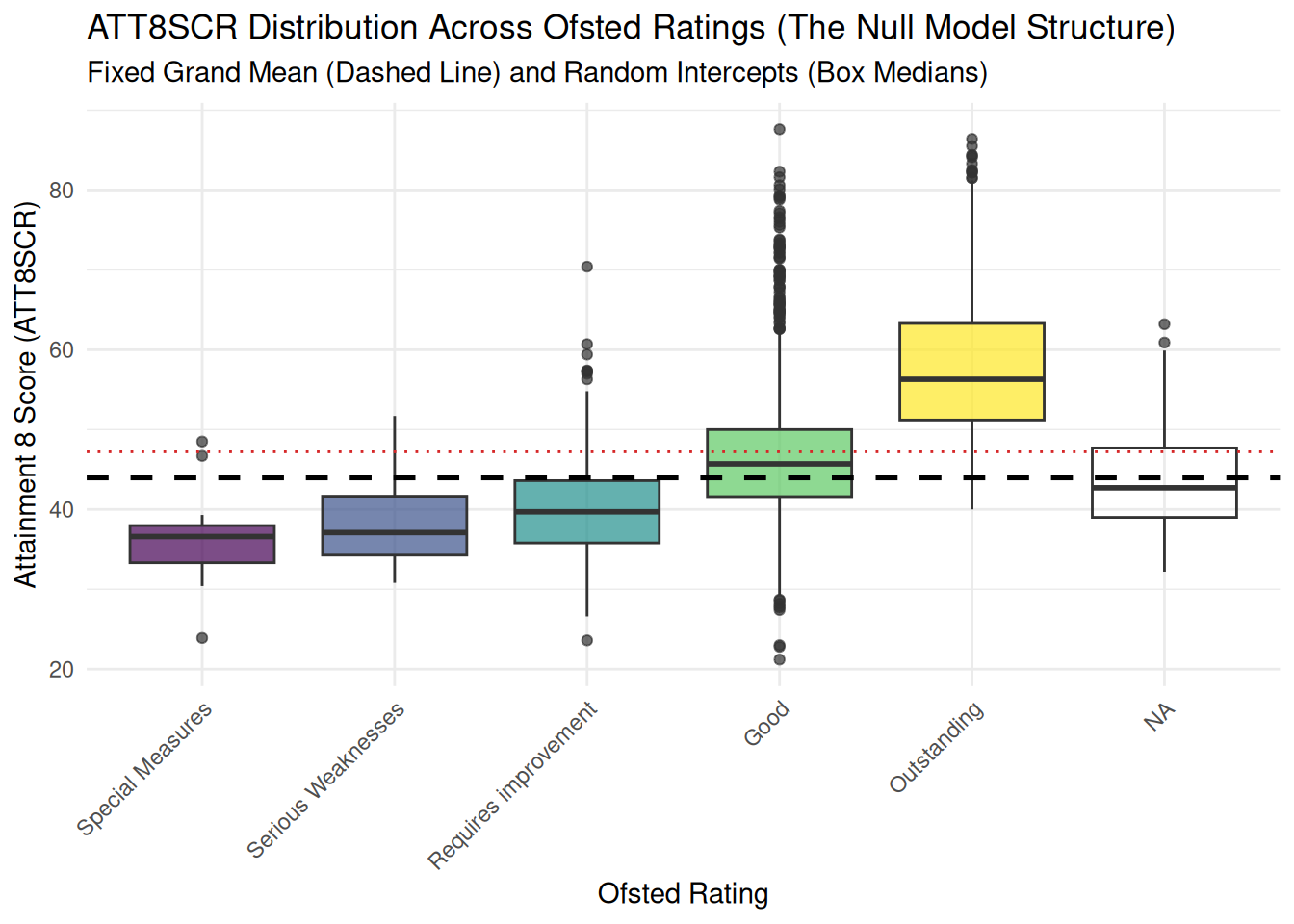
- Below is some code to visualise those groups as boxplots.
::: multicode
#### {width="30"}
```{python}
import pandas as pd
import numpy as np
import statsmodels.api as sm
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.lines import Line2D
sns.set_theme(style="whitegrid")
# --- Ensure OFSTEDRATING is clean and categorical ---
rating_order = ["Special Measures", "Serious Weaknesses", "Requires improvement", "Good", "Outstanding"]
england_filtered_clean["OFSTEDRATING"] = england_filtered_clean["OFSTEDRATING"].astype(str)
england_filtered_clean["OFSTEDRATING"] = pd.Categorical(
england_filtered_clean["OFSTEDRATING"],
categories=rating_order,
ordered=True
)
# --- Keep valid rows (ATT8SCR present and rating in our order) ---
df = england_filtered_clean.dropna(subset=["ATT8SCR"]).copy()
df = df[df["OFSTEDRATING"].isin(rating_order)]
# --- Fit MixedLM null model (random intercept by OFSTEDRATING) ---
mixed_model = sm.MixedLM.from_formula("ATT8SCR ~ 1", groups="OFSTEDRATING", data=df)
mixed_fit = mixed_model.fit(method="lbfgs")
# --- Global mean (fixed intercept) ---
global_mean = mixed_fit.params["Intercept"] if "Intercept" in mixed_fit.params.index else float(mixed_fit.params.iloc[0])
# --- Robust random-effects extraction ---
def get_re_value(effect):
# effect may be a pandas Series (with/without 'Intercept'), ndarray, or list
if isinstance(effect, pd.Series):
return float(effect["Intercept"]) if "Intercept" in effect.index else float(effect.iloc[0])
arr = np.asarray(effect).ravel()
return float(arr[0]) if arr.size > 0 else np.nan
random_effects = mixed_fit.random_effects # dict: {group_label: effect_vector}
group_effects = {str(k): get_re_value(v) for k, v in random_effects.items()}
ranef_df = pd.DataFrame({
"OFSTEDRATING": list(group_effects.keys()),
"ranef": list(group_effects.values())
}).sort_values("OFSTEDRATING")
# --- Actual intercepts = global mean + random effect ---
ranef_df["actual_intercept"] = global_mean + ranef_df["ranef"]
# --- Overall mean from raw data ---
overall_mean = df["ATT8SCR"].mean()
# --- Plot: boxplots + lines + actual intercept markers ---
fig, ax = plt.subplots(figsize=(10, 6))
sns.boxplot(data=df, x="OFSTEDRATING", y="ATT8SCR", palette="Set2", ax=ax)
# Reference lines
ax.axhline(global_mean, color="black", linestyle="--", linewidth=1)
ax.axhline(overall_mean, color="#d62728", linestyle=":", linewidth=1)
# Overlay actual intercepts (blue diamonds) per rating
sns.scatterplot(
data=ranef_df,
x="OFSTEDRATING", y="actual_intercept",
marker="D", s=100, color="blue",
ax=ax
)
# Titles & labels
ax.set_title("ATT8SCR Distribution Across Ofsted Ratings (Null Model Structure)", fontsize=14)
fig.suptitle("Dashed: Global Mean (MixedLM); Dotted: Overall Mean; Blue Diamonds: Actual Intercepts", fontsize=10)
ax.set_xlabel("Ofsted Rating")
ax.set_ylabel("Attainment 8 Score (ATT8SCR)")
plt.xticks(rotation=45)
# --- Manual legend handles (prevents 'Axes has no legend attached') ---
legend_handles = [
Line2D([0], [0], color='black', linestyle='--', linewidth=1, label='Global Mean (MixedLM)'),
Line2D([0], [0], color='#d62728', linestyle=':', linewidth=1, label='Overall Mean'),
Line2D([0], [0], marker='D', color='blue', linestyle='None', markersize=8, label='Actual Intercept (Global + RE)')
]
ax.legend(legend_handles, [h.get_label() for h in legend_handles],
loc="lower center", bbox_to_anchor=(0.5, -0.25), ncol=2, frameon=False)
plt.tight_layout()
plt.show()
```
#### {width="41" height="30"}
```{r}
#| message: false
#| warning: false
#| align: center
#| out-width: 80%
# 1. Load the necessary library for plotting
library(ggplot2)
library(lme4)
# --- Ensure OFSTEDRATING is an ordered factor for better plotting ---
# It appears you have six levels based on the OLS output, so we order them logically.
# Adjust the order if the true levels/baseline are different in your data.
rating_order <- c("Special Measures", "Serious Weaknesses", "Requires improvement", "Good", "Outstanding")
england_filtered_clean$OFSTEDRATING <- factor(
england_filtered_clean$OFSTEDRATING,
levels = rating_order,
ordered = TRUE
)
#Fit the model
lme1 <- lmer(ATT8SCR ~ 1 + (1 | OFSTEDRATING), data = england_filtered_clean)
# Extract the fixed-effect intercept and store in a variable
global_mean <- fixef(lme1)[1]
# Check the value
global_mean
## ## 1. Boxplot Visualization (Level 1 and Level 2 Variance) ## ##
boxplot_att8scr <- ggplot(england_filtered_clean, aes(x = OFSTEDRATING, y = ATT8SCR, fill = OFSTEDRATING)) +
geom_boxplot(alpha = 0.7) +
geom_hline(yintercept = global_mean, linetype = "dashed", color = "black", linewidth = 1) +
geom_hline(yintercept = mean(england_filtered_clean$ATT8SCR, na.rm = TRUE), linetype = "dotted", color = "#d62728", linewidth = 0.5) +
labs(title = "ATT8SCR Distribution Across Ofsted Ratings (The Null Model Structure)",
subtitle = "Fixed Grand Mean (Dashed Line) and Random Intercepts (Box Medians)",
x = "Ofsted Rating",
y = "Attainment 8 Score (ATT8SCR)") +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1), legend.position = "none")
print(boxplot_att8scr)
```
:::
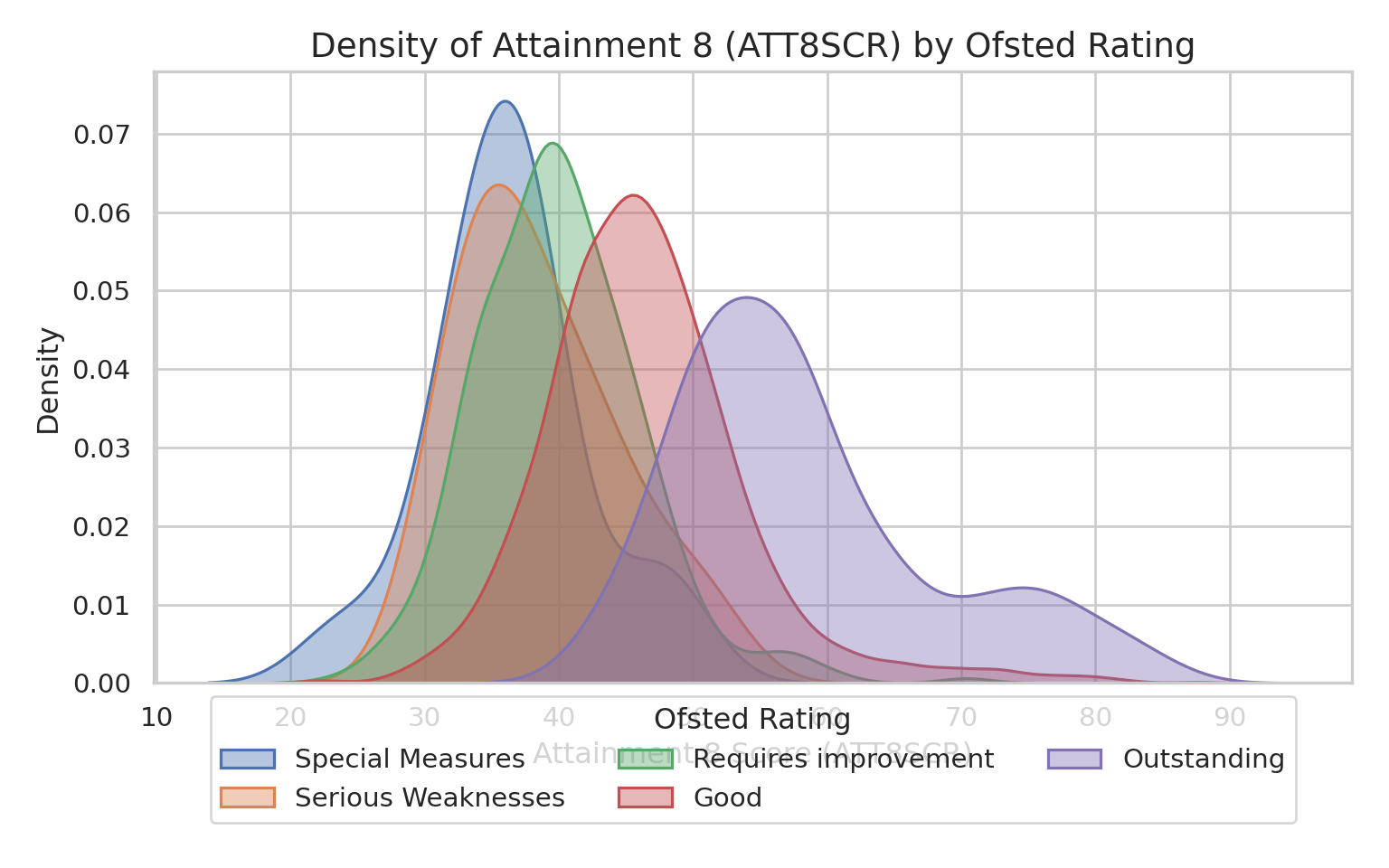
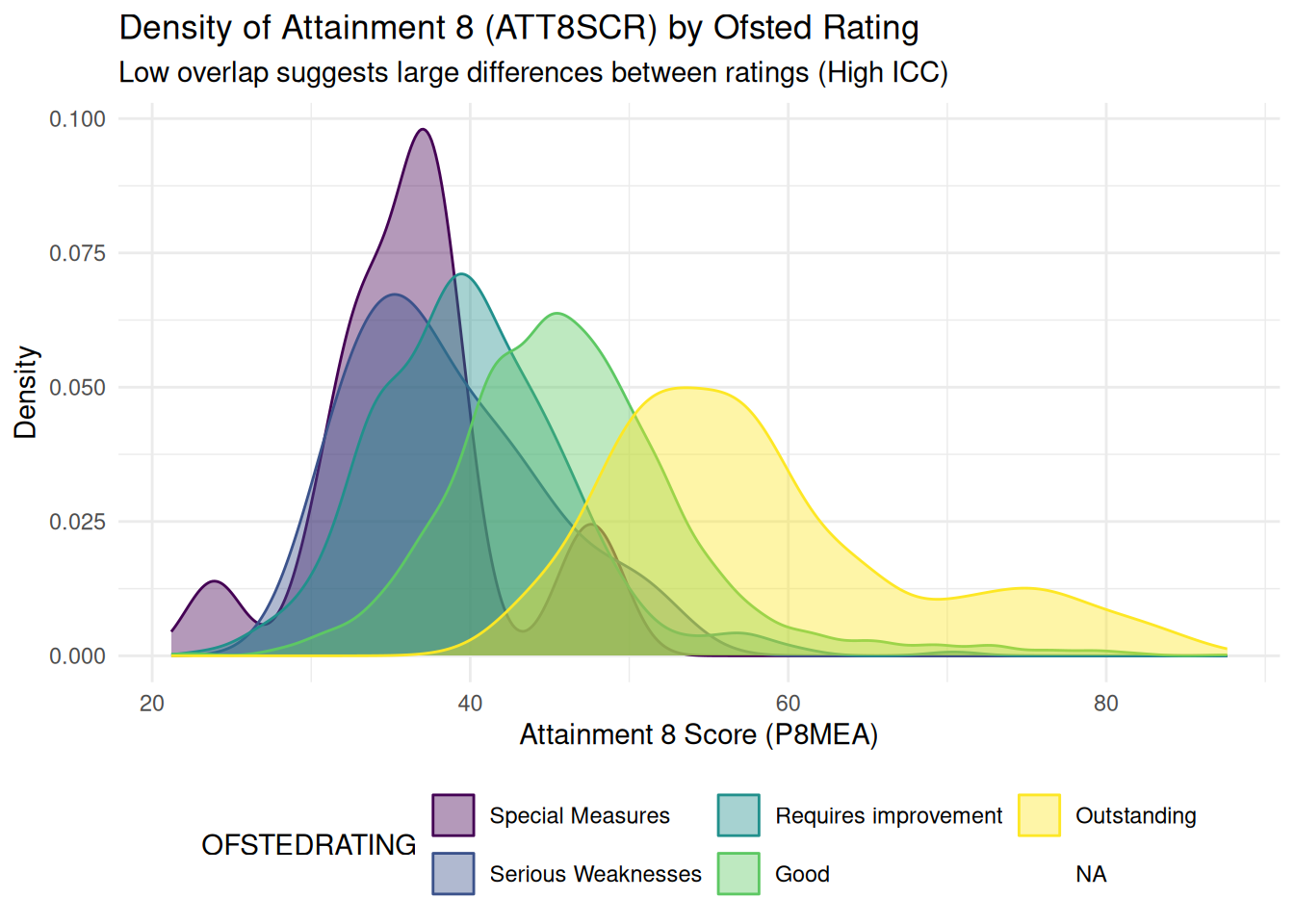
### The Interclass Correlation Coefficient - ICC
- Again, if you recall from the lecture, the ICC is simply the ratio
between the between the between group variance and the total
variance
- The higher the ICC (closer to 1), the more variance is due to
differences between groups (i.e. differences between the Ofsted
rating of schools) than within groups (i.e. between schools within
Ofsted rating bands)
- This can be judged visually with a stacked density plot:
- High ICC = distinct, non-overlapping humps
- Low ICC = density plots all overlapping on-top of each other
- **Remember** A high ICC is a strong indication that a multilevel /
mixed effects model is a suitable modelling strategy to pursue to
try and explain these group-level differences
::: multicode
#### {width="30"}
```{python}
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
sns.set_theme(style="whitegrid")
rating_order = ["Special Measures", "Serious Weaknesses", "Requires improvement", "Good", "Outstanding"]
england_filtered_clean["OFSTEDRATING"] = pd.Categorical(
england_filtered_clean["OFSTEDRATING"],
categories=rating_order,
ordered=True
)
plot_df = england_filtered_clean.dropna(subset=["ATT8SCR", "OFSTEDRATING"]).copy()
# Create figure and axis manually
fig, ax = plt.subplots(figsize=(8, 5))
# Plot KDE for each category
for rating in rating_order:
subset = plot_df[plot_df["OFSTEDRATING"] == rating]
sns.kdeplot(
data=subset,
x="ATT8SCR",
fill=True,
alpha=0.4,
linewidth=1.2,
ax=ax,
label=rating
)
# Titles & labels
ax.set_title("Density of Attainment 8 (ATT8SCR) by Ofsted Rating", fontsize=14)
ax.set_xlabel("Attainment 8 Score (ATT8SCR)")
ax.set_ylabel("Density")
ax.legend(title="Ofsted Rating", loc="lower center", bbox_to_anchor=(0.5, -0.25), ncol=3)
plt.tight_layout()
plt.show()
```
#### {width="41" height="30"}
```{r}
#| message: false
#| warning: false
#| align: center
#| out-width: 80%
## ## 2. Density Plot Visualization (Comparing the Two Variance Components) ## ##
# This plot visually shows how much the groups overlap (small overlap means high ICC).
density_att8scr <- ggplot(england_filtered_clean, aes(x = ATT8SCR, fill = OFSTEDRATING, color = OFSTEDRATING)) +
geom_density(alpha = 0.4) +
labs(
title = "Density of Attainment 8 (ATT8SCR) by Ofsted Rating",
subtitle = "Low overlap suggests large differences between ratings (High ICC)",
x = "Attainment 8 Score (P8MEA)",
y = "Density"
) +
theme_minimal() +
theme(legend.position = "bottom")
print(density_att8scr)
```
:::
## Running the Null Model and Calculating the Inter-Class Correlation Coefficient (ICC)
- You will also remember from the lecture, that we can run our null
model using standard statistical software. - In R, we use the `lmer`
package, whereas in Python for now we'll use `statsmodels`
- First run the model as below:
::: multicode
#### {width="30"}
```{python}
import pandas as pd
import statsmodels.api as sm
# Ensure OFSTEDRATING is categorical
rating_order = ["Special Measures", "Serious Weaknesses", "Requires improvement", "Good", "Outstanding"]
england_filtered_clean["OFSTEDRATING"] = pd.Categorical(
england_filtered_clean["OFSTEDRATING"],
categories=rating_order,
ordered=True
)
# Drop missing values for P8MEA and OFSTEDRATING
df = england_filtered_clean.dropna(subset=["ATT8SCR", "OFSTEDRATING"]).copy()
# Fit null model: random intercept for OFSTEDRATING
null_model = sm.MixedLM.from_formula("ATT8SCR ~ 1", groups="OFSTEDRATING", data=df)
null_fit = null_model.fit()
# Summary
print(null_fit.summary())
```
#### {width="41" height="30"}
```{r}
#| message: false
#| warning: false
#| align: center
#| out-width: 70%
library(lme4)
library(performance)
#lm1 <- lm(P8MEA ~ PTFSM6CLA1A + OFSTEDRATING, data = england_filtered_clean)
#summary(lm1)
lme1 <- lmer(ATT8SCR ~ 1 + (1|OFSTEDRATING), data = england_filtered_clean)
summary(lme1)
```
:::
- Once we have run the model we can calculate the ICC:
::: multicode
#### {width="30"}
```{python}
var_components = null_fit.cov_re.iloc[0, 0] # random intercept variance
resid_var = null_fit.scale # residual variance
icc = var_components / (var_components + resid_var)
print(f"ICC: {icc:.3f}")
```
#### {width="41" height="30"}
```{r}
#| message: false
#| warning: false
library(performance)
icc_value <- performance::icc(lme1, by_group = T)
print(icc_value)
```
:::
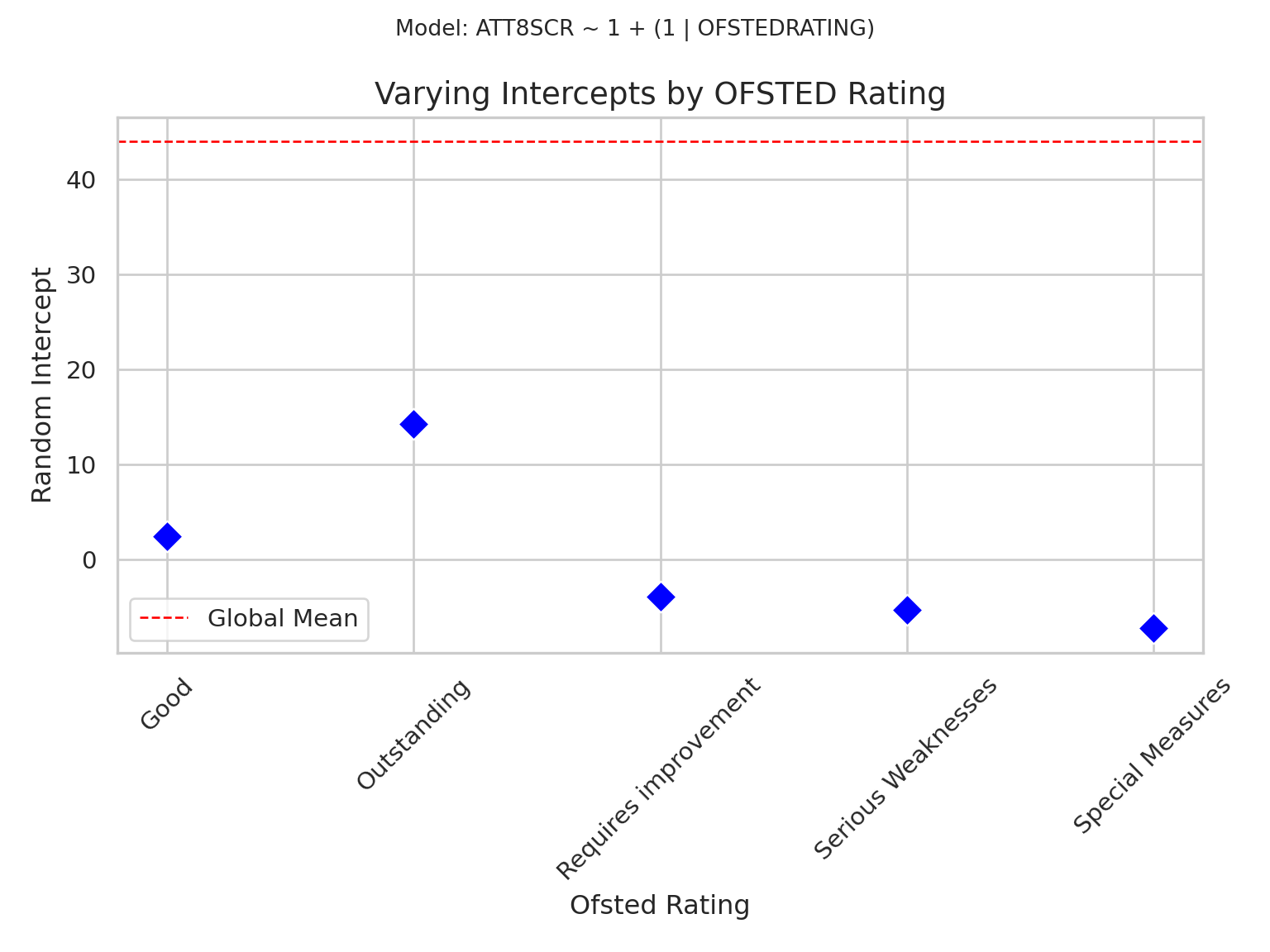
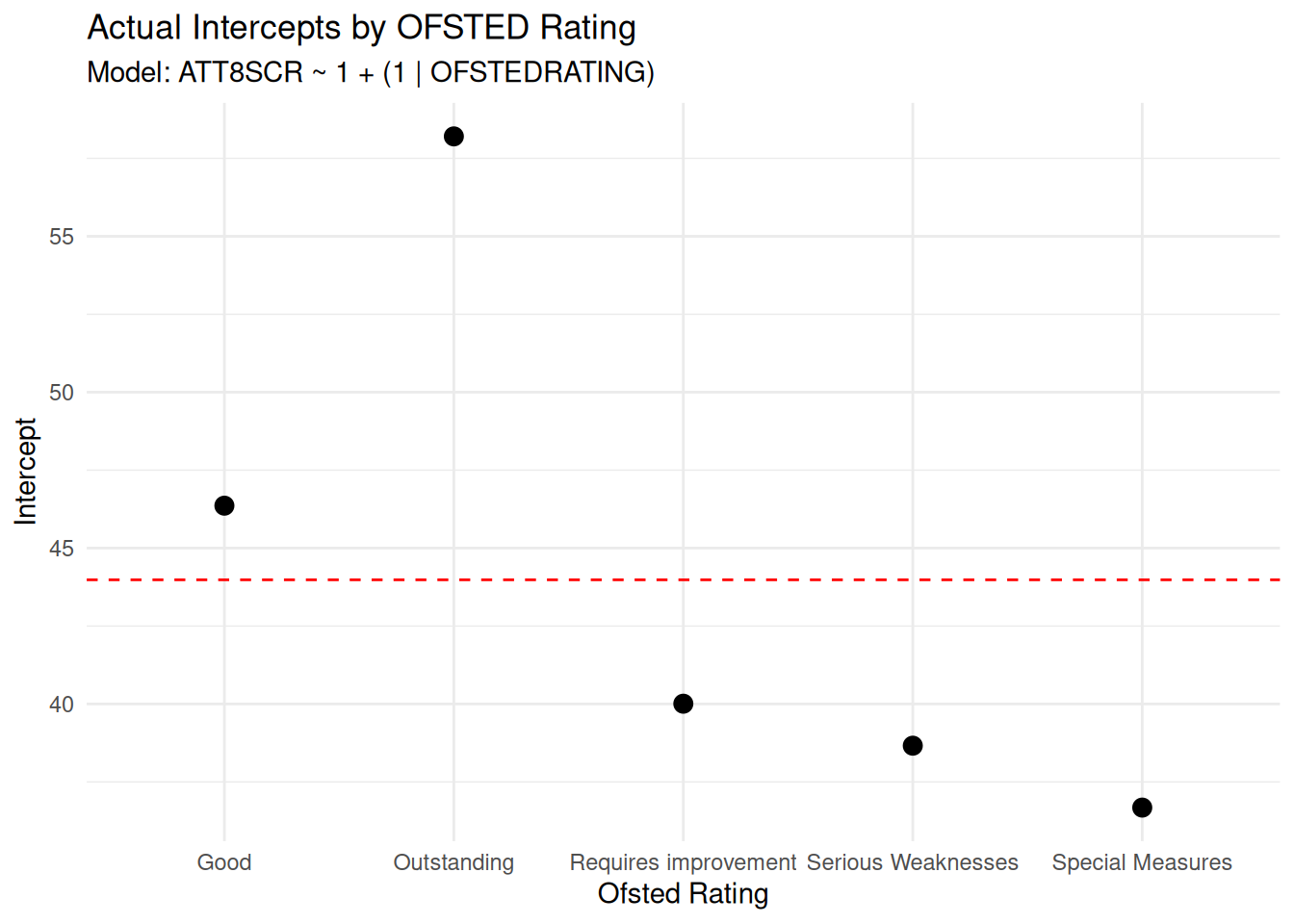
## Null Model Visualisation
- You will know by know that I am a big advovate of visualising
everything as we go, so here is a visualisation of the null model
which shows the different intercept values for the different group
- Remember, we can think of the intercept as the 'baseline' expected
for our dependent variable
::: multicode
#### {width="30"}
```{python}
import pandas as pd
import statsmodels.api as sm
import seaborn as sns
import matplotlib.pyplot as plt
# Ensure OFSTEDRATING is categorical
rating_order = ["Special Measures", "Serious Weaknesses", "Requires improvement", "Good", "Outstanding"]
england_filtered_clean["OFSTEDRATING"] = pd.Categorical(
england_filtered_clean["OFSTEDRATING"],
categories=rating_order,
ordered=True
)
# Drop missing values
df = england_filtered_clean.dropna(subset=["ATT8SCR", "OFSTEDRATING"]).copy()
# Fit null model: random intercept for OFSTEDRATING
null_model = sm.MixedLM.from_formula("ATT8SCR ~ 1", groups="OFSTEDRATING", data=df)
null_fit = null_model.fit()
# Extract global mean (fixed effect intercept)
global_mean = null_fit.params["Intercept"]
# Extract random effects for each group (take first value from each Series)
random_effects = null_fit.random_effects
ranef_df = pd.DataFrame({
"OFSTEDRATING": list(random_effects.keys()),
"Intercept": [float(effect.iloc[0]) for effect in random_effects.values()]
})
# Plot random intercepts
plt.figure(figsize=(8, 6))
sns.scatterplot(data=ranef_df, x="OFSTEDRATING", y="Intercept", s=100, color="blue", marker="D")
# Add dashed line for global mean
plt.axhline(global_mean, color="red", linestyle="--", linewidth=1, label="Global Mean")
# Titles and labels
plt.title("Varying Intercepts by OFSTED Rating", fontsize=14)
plt.suptitle("Model: ATT8SCR ~ 1 + (1 | OFSTEDRATING)", fontsize=10)
plt.xlabel("Ofsted Rating")
plt.ylabel("Random Intercept")
plt.xticks(rotation=45)
plt.legend()
plt.tight_layout()
plt.show()
```
#### {width="41" height="30"}
```{r}
#| message: false
#| warning: false
#| align: center
#| out-width: 80%
library(lme4)
library(ggplot2)
# Fit the model
lme1 <- lmer(ATT8SCR ~ 1 + (1|OFSTEDRATING), data = england_filtered_clean)
# Extract random effects
ranef_df <- as.data.frame(ranef(lme1)$OFSTEDRATING)
ranef_df$OFSTEDRATING <- rownames(ranef(lme1)$OFSTEDRATING)
ranef_df$actual_intercept <- ranef_df$`(Intercept)` + fixef(lme1)[1]
ggplot(ranef_df, aes(x = OFSTEDRATING, y = actual_intercept)) +
geom_point(size = 3) +
geom_hline(yintercept = fixef(lme1)[1], linetype = "dashed", color = "red") +
labs(title = "Actual Intercepts by OFSTED Rating",
subtitle = "Model: ATT8SCR ~ 1 + (1 | OFSTEDRATING)",
x = "Ofsted Rating",
y = "Intercept") +
theme_minimal()
```
:::
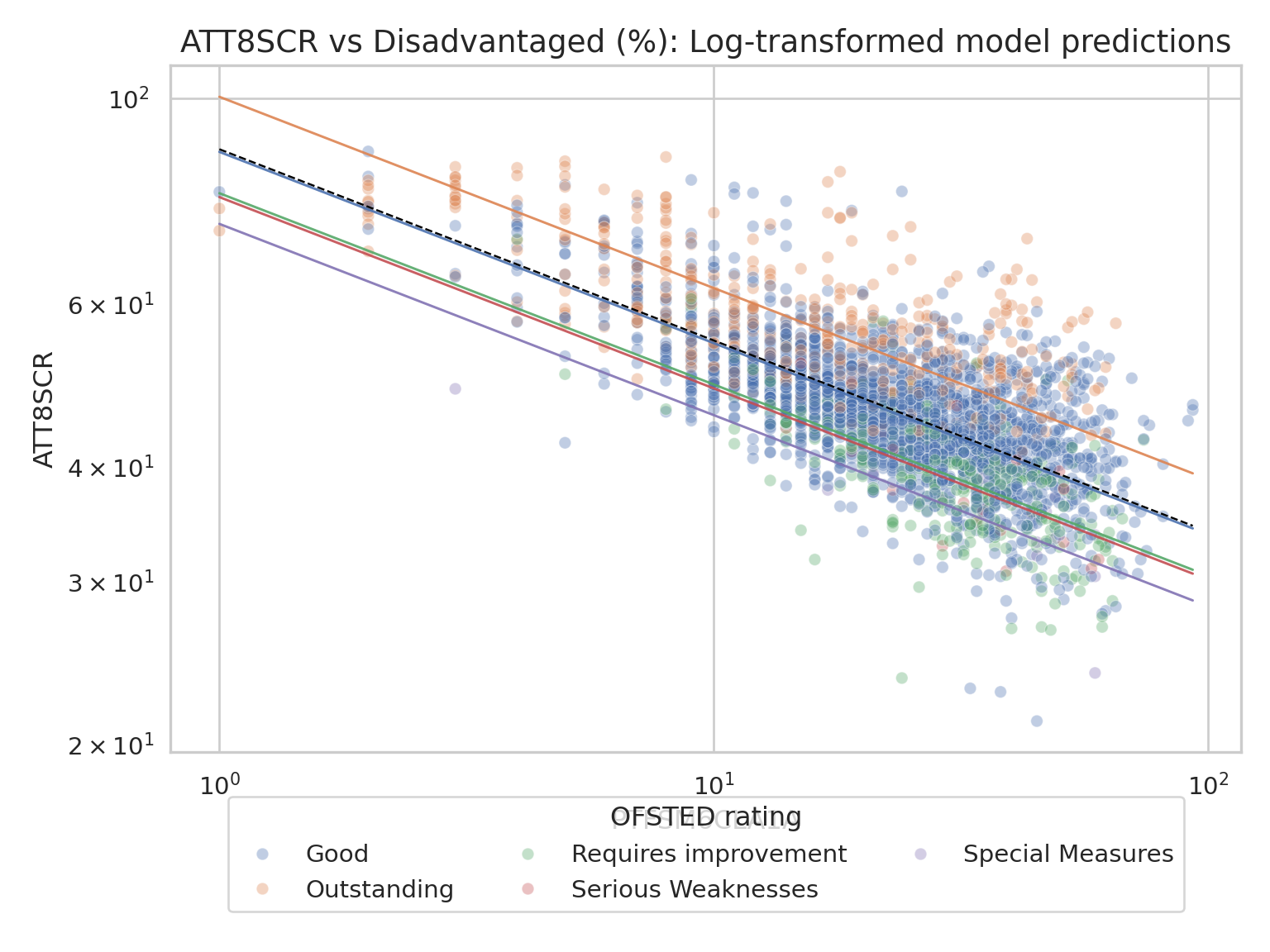
## Stage 2 - Adding an initial Fixed Effect
### Model with a Fixed Effect Predictor (random intercepts, fixed slopes)
- In the lecture you saw that after running the null model (assuming
the ICC indicates that a mixed effects model is recommended) the
next stage is to test that model by adding in a predictor variable
- You can use any predictor you like and once you get used to running
these models, you can even skip this step, but for now we are going
to build gradually and I am going to experiment with the %
disadvantaged variable
::: multicode
#### {width="30"}
```{python}
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import statsmodels.formula.api as smf
# 1. Clean data and check
df = england_filtered_clean.copy()
print("Original shape:", df.shape)
# Drop missing values
df = df.dropna(subset=["ATT8SCR", "PTFSM6CLA1A", "OFSTEDRATING"])
print("After dropna:", df.shape)
# Ensure correct types
df["OFSTEDRATING"] = df["OFSTEDRATING"].astype(str)
print("Groups:", df["OFSTEDRATING"].unique())
if df.empty:
raise ValueError("No data left after cleaning. Check column names and missing values.")
# 2. Apply log transformation
df["log_ATT8SCR"] = np.log(df["ATT8SCR"])
df["log_PTFSM6CLA1A"] = np.log(df["PTFSM6CLA1A"])
# 3. Fit simple linear model (fixed effect)
lm = smf.ols("log_ATT8SCR ~ log_PTFSM6CLA1A", data=df).fit()
fixed_intercept = lm.params["Intercept"]
slope = lm.params["log_PTFSM6CLA1A"]
print("Fixed effects:", lm.params)
# 4. Compute group-specific intercept adjustments
df["residual"] = df["log_ATT8SCR"] - (fixed_intercept + slope * df["log_PTFSM6CLA1A"])
group_adjustments = df.groupby("OFSTEDRATING")["residual"].mean().reset_index()
group_adjustments.rename(columns={"residual": "blup"}, inplace=True)
print("Group adjustments:", group_adjustments)
# 5. Prediction grid (log scale)
pop_range = np.linspace(df["log_PTFSM6CLA1A"].min(), df["log_PTFSM6CLA1A"].max(), 100)
# Build predictions for each group
pred_list = []
for _, row in group_adjustments.iterrows():
intercept_i = fixed_intercept + row["blup"]
pred_list.append(pd.DataFrame({
"log_PTFSM6CLA1A": pop_range,
"log_ATT8SCR": intercept_i + slope * pop_range,
"OFSTEDRATING": row["OFSTEDRATING"]
}))
pred_df = pd.concat(pred_list, ignore_index=True)
# Back-transform predictions to original scale
pred_df["PTFSM6CLA1A"] = np.exp(pred_df["log_PTFSM6CLA1A"])
pred_df["ATT8SCR"] = np.exp(pred_df["log_ATT8SCR"])
# Overall line
overall_df = pd.DataFrame({
"log_PTFSM6CLA1A": pop_range,
"log_ATT8SCR": fixed_intercept + slope * pop_range
})
overall_df["PTFSM6CLA1A"] = np.exp(overall_df["log_PTFSM6CLA1A"])
overall_df["ATT8SCR"] = np.exp(overall_df["log_ATT8SCR"])
# 6. Plot (original scale)
sns.set_theme(style="whitegrid")
fig, ax = plt.subplots(figsize=(8, 6))
# Raw points
sns.scatterplot(
data=df,
x="PTFSM6CLA1A", y="ATT8SCR", hue="OFSTEDRATING",
alpha=0.35, s=30, ax=ax
)
# Group-specific lines
sns.lineplot(
data=pred_df,
x="PTFSM6CLA1A", y="ATT8SCR", hue="OFSTEDRATING",
linewidth=1.1, alpha=0.9, ax=ax, legend=False
)
# Overall line
sns.lineplot(
data=overall_df,
x="PTFSM6CLA1A", y="ATT8SCR",
color="black", linestyle="--", linewidth=0.9, ax=ax
)
# Titles & labels
ax.set_title("ATT8SCR vs Disadvantaged (%): Log-transformed model predictions", fontsize=14)
ax.set_xlabel("PTFSM6CLA1A")
ax.set_ylabel("ATT8SCR")
ax.legend(title="OFSTED rating", loc="lower center", bbox_to_anchor=(0.5, -0.25), ncol=3)
ax.set_xscale("log")
ax.set_yscale("log")
plt.tight_layout()
plt.show()
```
#### {width="41" height="30"}
```{r}
#| message: false
#| warning: false
library(lme4)
library(ggplot2)
library(dplyr)
library(tibble)
# Fit the model
lme2 <- lmer(log(ATT8SCR) ~ log(PTFSM6CLA1A) + (1 | OFSTEDRATING), data = england_filtered_clean)
# Fixed effects
fixed_intercept <- fixef(lme2)[1]
slope <- fixef(lme2)[2]
# Random intercepts (BLUPs)
ranef_df <- ranef(lme2)$OFSTEDRATING %>%
as.data.frame() %>%
rownames_to_column("OFSTEDRATING") %>%
rename(blup = `(Intercept)`)
# Prediction grid across observed range (raw values)
pop_range <- seq(
min(england_filtered_clean$PTFSM6CLA1A, na.rm = TRUE),
max(england_filtered_clean$PTFSM6CLA1A, na.rm = TRUE),
length.out = 100
)
# Lines for each OFSTED group: apply log in prediction
pred_df <- do.call(rbind, lapply(seq_len(nrow(ranef_df)), function(i) {
intercept_i <- fixed_intercept + ranef_df$blup[i]
data.frame(
PTFSM6CLA1A = pop_range,
ATT8SCR = exp(intercept_i + slope * log(pop_range)), # back-transform to original scale
OFSTEDRATING = ranef_df$OFSTEDRATING[i],
stringsAsFactors = FALSE
)
}))
# Overall fixed-effects line
overall_df <- data.frame(
PTFSM6CLA1A = pop_range,
ATT8SCR = exp(fixed_intercept + slope * log(pop_range)) # back-transform
)
# Plot: raw points and fitted lines
ggplot() +
geom_point(
data = england_filtered_clean,
aes(x = log(PTFSM6CLA1A), y = log(ATT8SCR), color = OFSTEDRATING),
alpha = 0.35, size = 1.2, na.rm = TRUE
) +
geom_line(
data = pred_df,
aes(x = log(PTFSM6CLA1A), y = log(ATT8SCR), color = OFSTEDRATING),
linewidth = 1.1, alpha = 0.9
) +
geom_line(
data = overall_df,
aes(x = log(PTFSM6CLA1A), y = log(ATT8SCR)),
color = "black", linetype = "dashed", linewidth = 0.9
) +
scale_color_brewer(palette = "Set2") +
labs(
title = "log(ATT8SCR) vs log(Disadvantaged): Raw data and fitted lines",
subtitle = "Model: log(ATT8SCR) ~ log(PTFSM6CLA1A) + (1 | OFSTEDRATING)",
x = "log(PTFSM6CLA1A)",
y = "log(ATT8SCR)",
color = "OFSTED rating"
) +
theme_minimal(base_size = 12)
```
:::
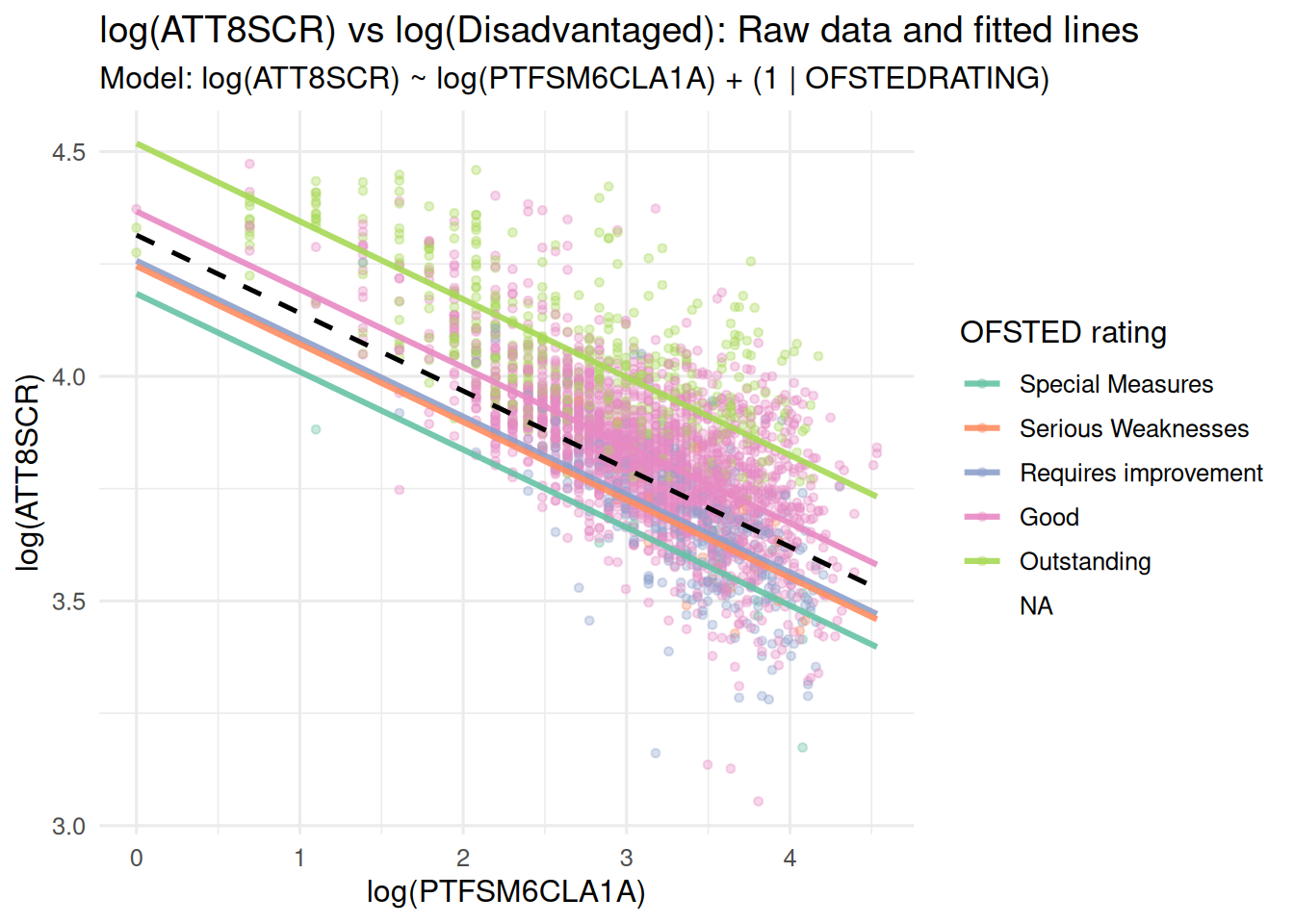
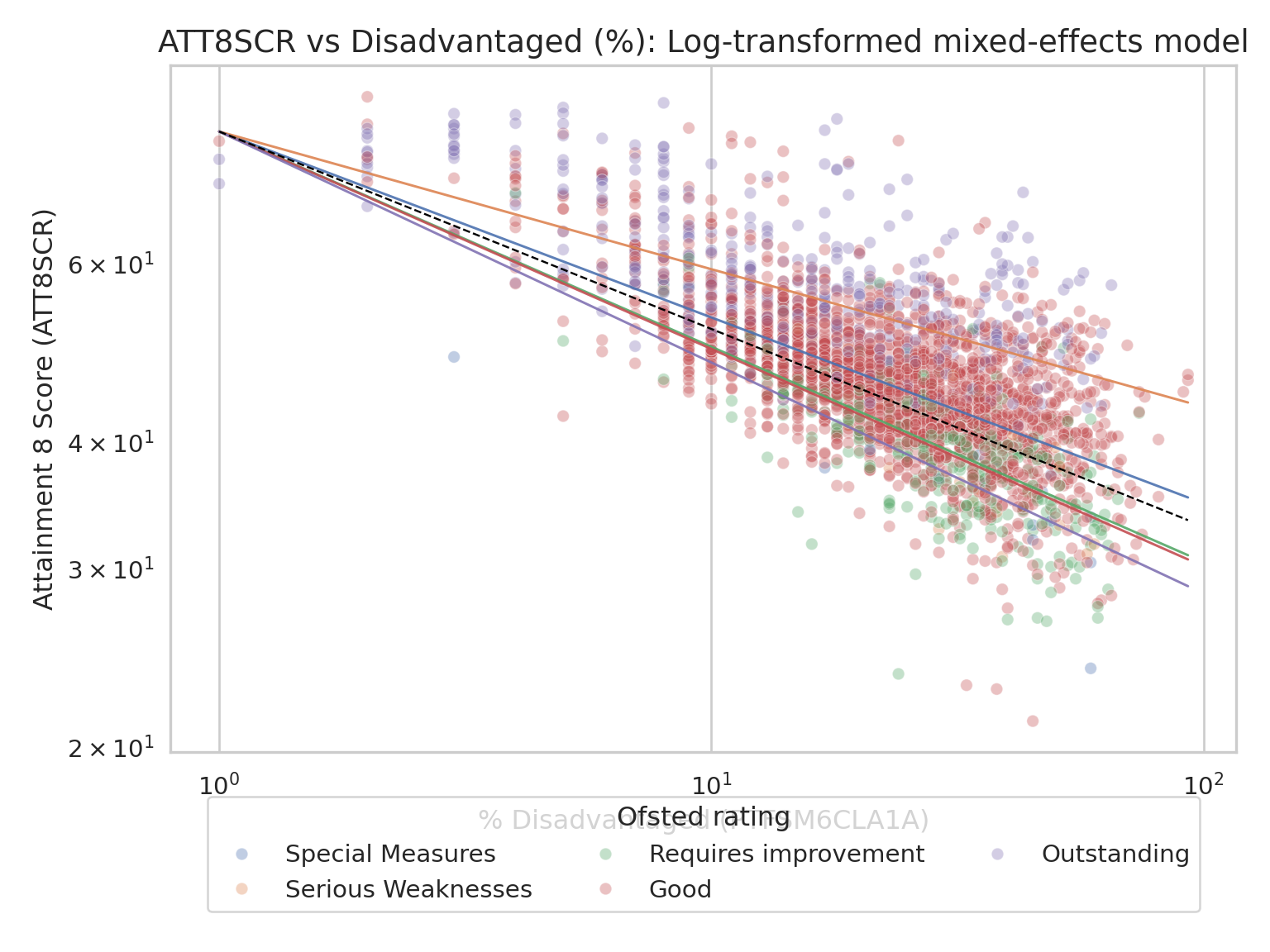
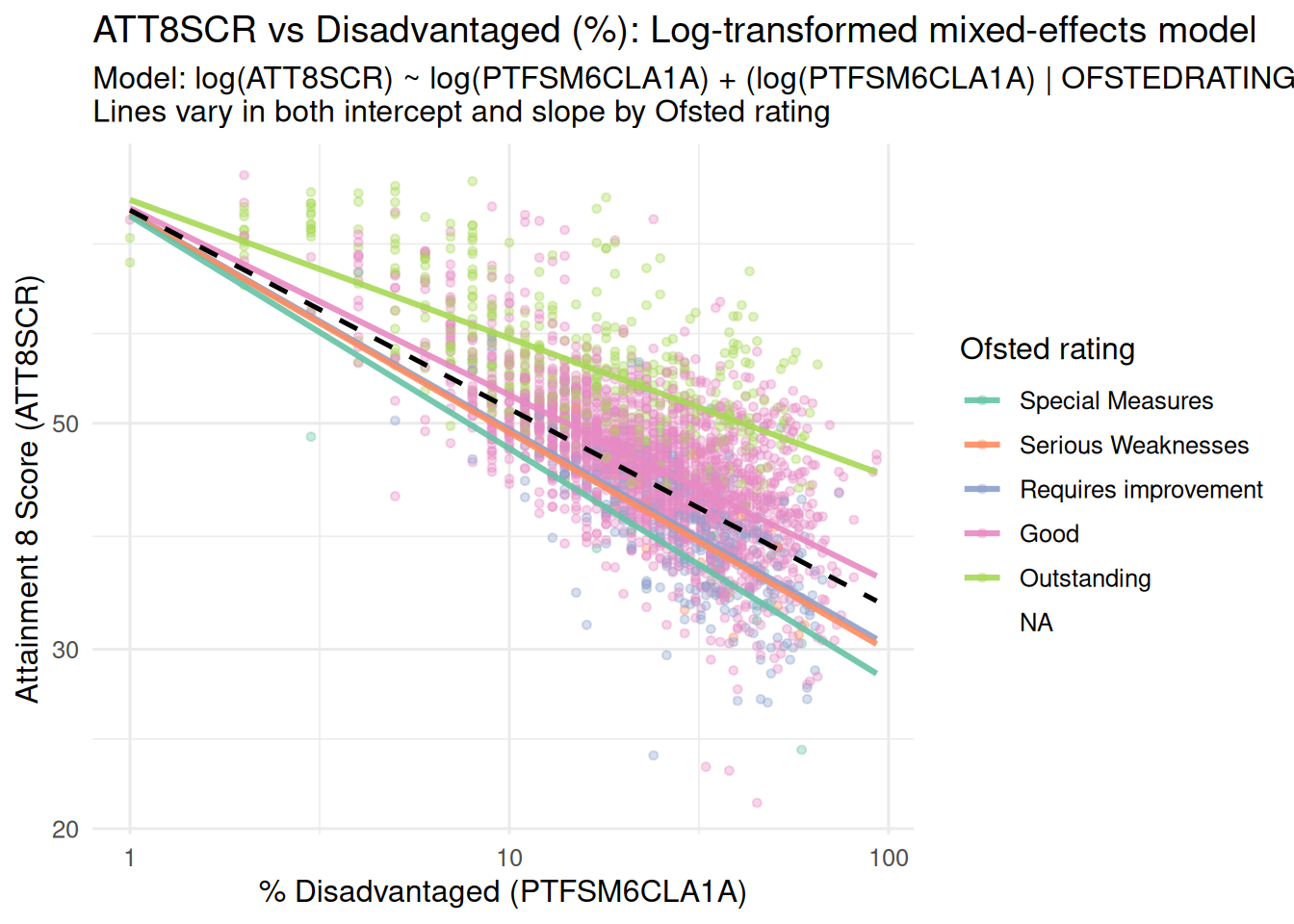
### Random Intercepts and Random Slopes Model
- And again, in the lecture we saw that the next stage once you have
allowed your intercepts to vary, is to allow your slopes to vary as
well
- Here's the visualisation of those varying slopes and intercepts:
::: multicode
#### {width="30"}
```{python}
import pandas as pd
import numpy as np
import statsmodels.formula.api as smf
import matplotlib.pyplot as plt
import seaborn as sns
df = england_filtered_clean.copy()
df["log_ATT8SCR"] = np.log(df["ATT8SCR"])
df["log_PTFSM6CLA1A"] = np.log(df["PTFSM6CLA1A"])
# Drop rows with missing values in any relevant column
df = df.dropna(subset=["log_ATT8SCR", "log_PTFSM6CLA1A", "OFSTEDRATING"])
# Fit mixed model with random intercepts and slopes
model = smf.mixedlm(
"log_ATT8SCR ~ log_PTFSM6CLA1A",
df,
groups=df["OFSTEDRATING"],
re_formula="~log_PTFSM6CLA1A"
)
lme_rs = model.fit(reml=False, method="lbfgs")
# Fixed effects
fixed_intercept = lme_rs.fe_params["Intercept"]
fixed_slope = lme_rs.fe_params["log_PTFSM6CLA1A"]
# Random effects per group (intercept and slope deviations)
ranef = lme_rs.random_effects
re_df = pd.DataFrame([
{
"OFSTEDRATING": g,
"b0": v.get("Intercept", v.get("0", 0)), # intercept deviation
"b1": v.get("log_PTFSM6CLA1A", 0) # slope deviation
}
for g, v in ranef.items()
])
# Prediction grid (log scale)
x_range_log = np.linspace(np.log(df["PTFSM6CLA1A"].min()), np.log(df["PTFSM6CLA1A"].max()), 100)
# Build predictions for each group
pred_list = []
for _, row in re_df.iterrows():
intercept_i = fixed_intercept + row["b0"]
slope_i = fixed_slope + row["b1"]
pred_list.append(pd.DataFrame({
"OFSTEDRATING": row["OFSTEDRATING"],
"PTFSM6CLA1A": np.exp(x_range_log),
"ATT8SCR": np.exp(intercept_i + slope_i * x_range_log)
}))
pred_df = pd.concat(pred_list)
# Overall fixed-effects line
overall_df = pd.DataFrame({
"PTFSM6CLA1A": np.exp(x_range_log),
"ATT8SCR": np.exp(fixed_intercept + fixed_slope * x_range_log)
})
# Plot
sns.set_theme(style="whitegrid")
fig, ax = plt.subplots(figsize=(8, 6))
sns.scatterplot(data=df, x="PTFSM6CLA1A", y="ATT8SCR", hue="OFSTEDRATING", alpha=0.35, s=30, ax=ax)
sns.lineplot(data=pred_df, x="PTFSM6CLA1A", y="ATT8SCR", hue="OFSTEDRATING", linewidth=1.1, alpha=0.9, ax=ax, legend=False)
sns.lineplot(data=overall_df, x="PTFSM6CLA1A", y="ATT8SCR", color="black", linestyle="--", linewidth=0.9, ax=ax)
ax.set_xscale("log")
ax.set_yscale("log")
ax.set_title("ATT8SCR vs Disadvantaged (%): Log-transformed mixed-effects model", fontsize=14)
ax.set_xlabel("% Disadvantaged (PTFSM6CLA1A)")
ax.set_ylabel("Attainment 8 Score (ATT8SCR)")
ax.legend(title="Ofsted rating", loc="lower center", bbox_to_anchor=(0.5, -0.25), ncol=3)
plt.tight_layout()
plt.show()
```
#### {width="41" height="30"}
```{r}
#| message: false
#| warning: false
# Packages
library(lme4)
library(ggplot2)
library(dplyr)
library(tibble)
# --- Prepare log-transformed variables
england_filtered_clean <- england_filtered_clean %>%
mutate(
log_ATT8SCR = log(ATT8SCR),
log_PTFSM6CLA1A = log(PTFSM6CLA1A)
)
# --- Fit random intercept + random slope model
lme_rs <- lmer(log_ATT8SCR ~ log_PTFSM6CLA1A + (log_PTFSM6CLA1A | OFSTEDRATING),
data = england_filtered_clean)
# Fixed effects
fixed_intercept <- fixef(lme_rs)[["(Intercept)"]]
fixed_slope <- fixef(lme_rs)[["log_PTFSM6CLA1A"]]
# Random effects (BLUPs) per group: intercept and slope
re_df <- ranef(lme_rs)$OFSTEDRATING %>%
rownames_to_column("OFSTEDRATING") %>%
rename(
b0 = `(Intercept)`, # random intercept deviation
b1 = `log_PTFSM6CLA1A` # random slope deviation
)
# Prediction grid across observed range (log scale)
x_range_log <- seq(
min(log(england_filtered_clean$PTFSM6CLA1A), na.rm = TRUE),
max(log(england_filtered_clean$PTFSM6CLA1A), na.rm = TRUE),
length.out = 100
)
# Build per-group prediction lines (varying intercepts & slopes)
pred_df <- do.call(rbind, lapply(seq_len(nrow(re_df)), function(i) {
intercept_i <- fixed_intercept + re_df$b0[i]
slope_i <- fixed_slope + re_df$b1[i]
data.frame(
OFSTEDRATING = re_df$OFSTEDRATING[i],
PTFSM6CLA1A = exp(x_range_log),
ATT8SCR = exp(intercept_i + slope_i * x_range_log),
stringsAsFactors = FALSE
)
}))
# Overall fixed-effects line
overall_df <- data.frame(
PTFSM6CLA1A = exp(x_range_log),
ATT8SCR = exp(fixed_intercept + fixed_slope * x_range_log)
)
# --- Plot: raw data + group-specific fitted lines
ggplot() +
# Raw points
geom_point(
data = england_filtered_clean,
aes(x = PTFSM6CLA1A, y = ATT8SCR, color = OFSTEDRATING),
alpha = 0.35, size = 1.2, na.rm = TRUE
) +
# Group-specific lines
geom_line(
data = pred_df,
aes(x = PTFSM6CLA1A, y = ATT8SCR, color = OFSTEDRATING),
linewidth = 1.1, alpha = 0.9
) +
# Overall line
geom_line(
data = overall_df,
aes(x = PTFSM6CLA1A, y = ATT8SCR),
color = "black", linetype = "dashed", linewidth = 0.9
) +
scale_color_brewer(palette = "Set2", name = "Ofsted rating") +
scale_x_log10() +
scale_y_log10() +
labs(
title = "ATT8SCR vs Disadvantaged (%): Log-transformed mixed-effects model",
subtitle = "Model: log(ATT8SCR) ~ log(PTFSM6CLA1A) + (log(PTFSM6CLA1A) | OFSTEDRATING)\nLines vary in both intercept and slope by Ofsted rating",
x = "% Disadvantaged (PTFSM6CLA1A)",
y = "Attainment 8 Score (ATT8SCR)"
) +
theme_minimal(base_size = 12)
```
:::
### Fitting a Basic Random Effect and Random Slope Model
- And now we can also fit the model using statistical software
::: multicode
#### {width="30"}
```{python}
import pandas as pd
import statsmodels.formula.api as smf
df_clean = df.dropna(subset=["log_ATT8SCR", "log_PTFSM6CLA1A", "OFSTEDRATING"])
# Example
model = smf.mixedlm(
"log_ATT8SCR ~ log_PTFSM6CLA1A",
data=df_clean,
groups=df_clean["OFSTEDRATING"],
re_formula="~log_PTFSM6CLA1A"
)
result = model.fit()
print(result.summary())
```
#### {width="41" height="30"}
```{r}
lme3 <- lmer(log(ATT8SCR) ~ log(PTFSM6CLA1A) + (log(PTFSM6CLA1A)|OFSTEDRATING), data = england_filtered_clean)
summary(lme3)
```
:::
### Interpretation
**NB - this applies to these models above, you will have different
interpretations depending on the variables you have chosen**
Here we have fitted a basic random intercept and random slope model
using comparable methods in R and Python.
#### Fixed Effects
Intercept (≈ 4.39) This is the expected value of log(ATT8SCR) when
log(PTFSM6CLA1A) is zero (i.e., when the predictor is at its baseline).
In practical terms, it’s the average log score for schools at the
reference level of the predictor.
Slope for log(PTFSM6CLA1A) (≈ −0.195) For each 1-unit increase in the
log of PTFSM6CLA1A, the log of ATT8SCR decreases by about 0.195. Since
both variables are logged, this is roughly an elasticity: higher
PTFSM6CLA1A is associated with lower ATT8SCR.
#### Random Effects
OFSTEDRATING intercept variance (≈ 0.00021) There’s very little
variation in baseline performance across OFSTED categories after
accounting for the fixed effect.
Random slope variance (≈ 0.00138) The effect of log(PTFSM6CLA1A) barely
varies by OFSTED category—almost zero. The correlation between intercept
and slope is estimated as 1.00, which is why you see the “singular fit”
warning (R) and Convergence Warnings in Python: the model is on the
boundary, meaning the random slope is not really supported by the data.
Residual variance (≈ 0.014) This is the within-group variability not
explained by the model.
## Experimenting with your own linear mixed effects models
At this point, this is about all you need to know, for now, to run your
own linear mixed effects model.
In general you might find it's a bit easier to run these models in R
using the `lme4` package, however, there are Python Packages that allow
you to run linear mixed effects models using R-like syntax like
`Pymer4`.
Please experiment - I was running into problems with `Pymer4` in this
practical, so you might need to persevere a bit if you want to persist
on the Python track.
`Pymer4` - <https://eshinjolly.com/pymer4/>
`lme4` - <https://github.com/lme4/lme4/>
I didn't touch on the Bayesian versions of these models either (a world
that would have taken us many more weeks to get into!). But you can run
Bayesian Linear Mixed Effects models too and there are packages in both
R and Python to do this.
In Python you have `bambi` - <https://bambinos.github.io/bambi/>
In R you have `RStan` - <https://mc-stan.org/rstan/index.html>
If you remember from the lecture, there are a range of variations on the
basic model which may or may not be appropriate in different situations.
I'll reproduce the table below:
| Element | Meaning | Sample Equation |
|------------------|------------------------------|------------------------|
| `(1|g)` | Null Model or Varying Intercept Model | `P8 ~ 1 + (1 | OFSTEDRATING)` |
| `(1|g1/g2)` | Multilevel Model with intercept varying within g1 and for g2 within g1 | `P8 ~ 1 + (1 | LocalAuthority/Region)` |
| `(1|g1) + (1|g2)` | Intercept varying among g1 and g2 | `P8 ~ 1 + (1 | OFSTEDRATING) + (1 | Region)` |
| `(0+x|g)` | Varying Slope Model | `P8 ~ Disadvantage + (0 + Disadvantage | OFSTEDRATING)` |
| `x + (x|g)` | Varying and correlated intercepts and slopes | `P8 ~ Disadvantage + (Disadvantage | OFSTEDRATING)` |
| `x + (x||g)` | Uncorrelated Intercepts and Slopes | `P8 ~ Disadvantage + (Disadvantage || OFSTEDRATING)` |
- You might want to experiment with only one or two of these linear
mixed effects models. Below you will see I have incorporated both
multilevel (local authorities nested within regions) and varying
intercepts. There are almost infinite combinations.
### Final Task
- You should try and extend your final multiple regression showstopper
model to incorporate one or more logical random effect. What can you
say about the random effects in your model?
- I have played around with a few additional variables too, you don't
have to do this, but you can if you wish.
```{r}
#| eval: false
#| include: false
england_filtered_clean <- england_filtered_clean %>%
filter(PTFSM6CLA1A > 0, PERCTOT > 0, PNUMEAL > 0)
# Before fitting the model:
contrasts(england_filtered_clean$OFSTEDRATING) <- contr.treatment(levels(england_filtered_clean$OFSTEDRATING))
lme9 <- lmer(log(ATT8SCR) ~ log(PTFSM6CLA1A) + log(PERCTOT) + log(PNUMEAL) + PTPRIORLO + ADMPOL_PT + (1|OFSTEDRATING) + (1|gor_name/LANAME), data = england_filtered_clean, na.action = na.exclude)
summary(lme9)
```
```{r}
#| include: false
library(lme4)
library(dplyr)
library(ggplot2)
library(ggrepel)
# Filter and prepare data
england_filtered_clean <- england_filtered_clean %>%
filter(PTFSM6CLA1A > 0, PERCTOT > 0, PNUMEAL > 0)
# Fit mixed-effects model
lme9 <- lmer(
log(ATT8SCR) ~ log(PTFSM6CLA1A) + log(PERCTOT) + log(PNUMEAL) +
PTPRIORLO + ADMPOL_PT + (1 | OFSTEDRATING) + (1 | gor_name/LANAME),
data = england_filtered_clean,
na.action = na.exclude
)
# Add fitted values (back-transform to original scale)
england_filtered_clean <- england_filtered_clean %>%
mutate(fitted_ATT8SCR = exp(fitted(lme9)))
# Compute R²
r2 <- cor(england_filtered_clean$ATT8SCR, england_filtered_clean$fitted_ATT8SCR, use = "complete.obs")^2
# Identify Brighton schools
brighton_schools <- england_filtered_clean %>%
filter(LANAME == "Brighton and Hove")
# Plot
ggplot(england_filtered_clean, aes(x = fitted_ATT8SCR, y = ATT8SCR)) +
geom_point(color = "grey70", alpha = 0.6) +
geom_point(data = brighton_schools, color = "orange", size = 2) +
geom_text_repel(data = brighton_schools,
aes(label = SCHNAME.x),
color = "black",
size = 3,
max.overlaps = 20) +
geom_abline(slope = 1, intercept = 0, linetype = "dashed", color = "black") +
labs(
title = paste0("Observed vs Fitted ATT8SCR (R² = ", round(r2, 3), ")"),
x = "Fitted ATT8SCR",
y = "Observed ATT8SCR"
) +
theme_minimal()
```
```{r}
#| eval: false
#| include: false
lme10 <- lmer(
log(ATT8SCR) ~ log(PTFSM6CLA1A) + log(PERCTOT) + log(PNUMEAL) +
PTPRIORLO + ADMPOL_PT + gorard_segregation +
(1 | OFSTEDRATING) + (1 | gor_name/LANAME),
data = england_filtered_clean,
na.action = na.exclude
)
summary(lme10)
```
```{r}
#| include: false
library(dplyr)
# Filter out rows with zeros or negatives in variables to be logged
england_filtered_clean <- england_filtered_clean %>%
filter(
ATT8SCR > 0,
PTFSM6CLA1A > 0,
PERCTOT > 0,
PNUMEAL > 0,
average_number_of_days_taken.x > 0,
remained_in_the_same_school > 0
)
# Log-transform variables
england_filtered_clean <- england_filtered_clean %>%
mutate(
log_ATT8SCR = log(ATT8SCR),
log_days_taken = log(average_number_of_days_taken.x),
log_remained = log(remained_in_the_same_school)
)
england_filtered_clean <- england_filtered_clean %>%
mutate(
retention_rate <- remained_in_the_same_school / teacher_fte_in_census_year
)
# Fit the linear model
england_model_test <- lm(
log_ATT8SCR ~ log(PTFSM6CLA1A) + log(PERCTOT) + log(PNUMEAL) +
OFSTEDRATING + gor_name + PTPRIORLO + ADMPOL_PT + gorard_segregation +
log_remained + teachers_on_leadership_pay_range_percent + log_days_taken,
data = england_filtered_clean,
na.action = na.exclude
)
# View summary
summary(england_model_test)
```
#### My final mixed effects model
::: multicode
#### {width="30"}
```{python}
#Sorry, I really tried to get this working properly with pymer, but I couldn't - too much pain.
# Try the R version instead, or, if you manage to get as far as getting pymer4 to work, you can translate my model below into pymer4 syntax (as it uses R under the hood)
```
#### {width="41" height="30"}
```{r}
lme11 <- lmer(
log(ATT8SCR) ~ log(PTFSM6CLA1A) + log(PERCTOT) + log(PNUMEAL) +
PTPRIORLO + ADMPOL_PT + gorard_segregation +
log(remained_in_the_same_school) + teachers_on_leadership_pay_range_percent + log(average_number_of_days_taken.x) +
(1 | OFSTEDRATING) + (1 | gor_name/LANAME),
data = england_filtered_clean,
na.action = na.exclude
)
summary(lme11)
```
:::
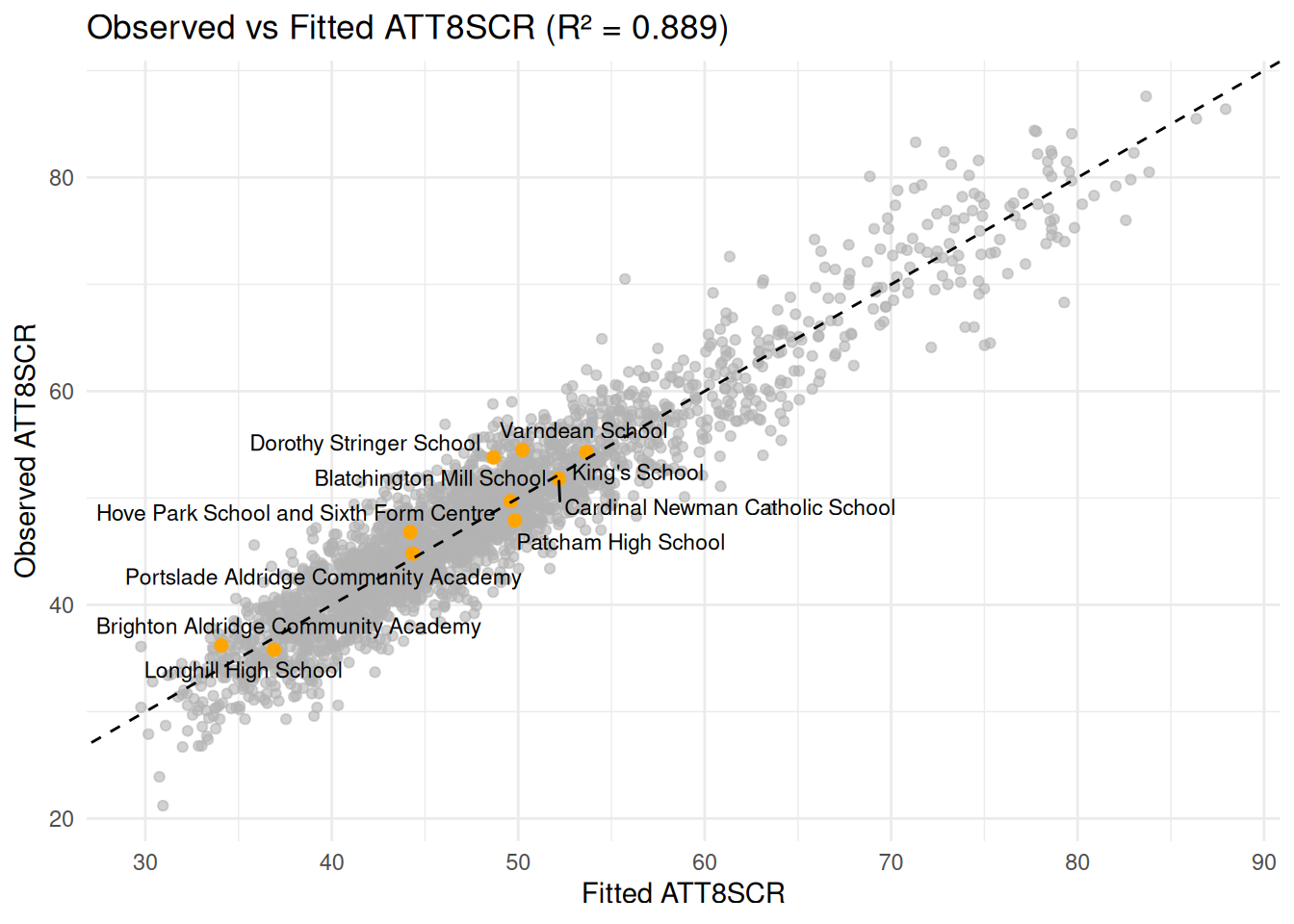
#### Fitted Values vs Observed Attainment 8 (how good is the model?)
- Here I have overlaid the Brighton Schools again - anything above the
line (a positive residual) shows the school is doing better than
expected, given the variables we have used to predict attainment.
Anything below, the school is performing worse than expected.
```{r}
# Add fitted values (back-transform to original scale)
england_filtered_clean <- england_filtered_clean %>%
mutate(fitted_ATT8SCR = exp(fitted(lme11)))
# Compute R²
r2 <- cor(england_filtered_clean$ATT8SCR, england_filtered_clean$fitted_ATT8SCR, use = "complete.obs")^2
# Identify Brighton schools
brighton_schools <- england_filtered_clean %>%
filter(LANAME == "Brighton and Hove")
# Plot
ggplot(england_filtered_clean, aes(x = fitted_ATT8SCR, y = ATT8SCR)) +
geom_point(color = "grey70", alpha = 0.6) +
geom_point(data = brighton_schools, color = "orange", size = 2) +
geom_text_repel(data = brighton_schools,
aes(label = SCHNAME.x),
color = "black",
size = 3,
max.overlaps = 20) +
geom_abline(slope = 1, intercept = 0, linetype = "dashed", color = "black") +
labs(
title = paste0("Observed vs Fitted ATT8SCR (R² = ", round(r2, 3), ")"),
x = "Fitted ATT8SCR",
y = "Observed ATT8SCR"
) +
theme_minimal()
```
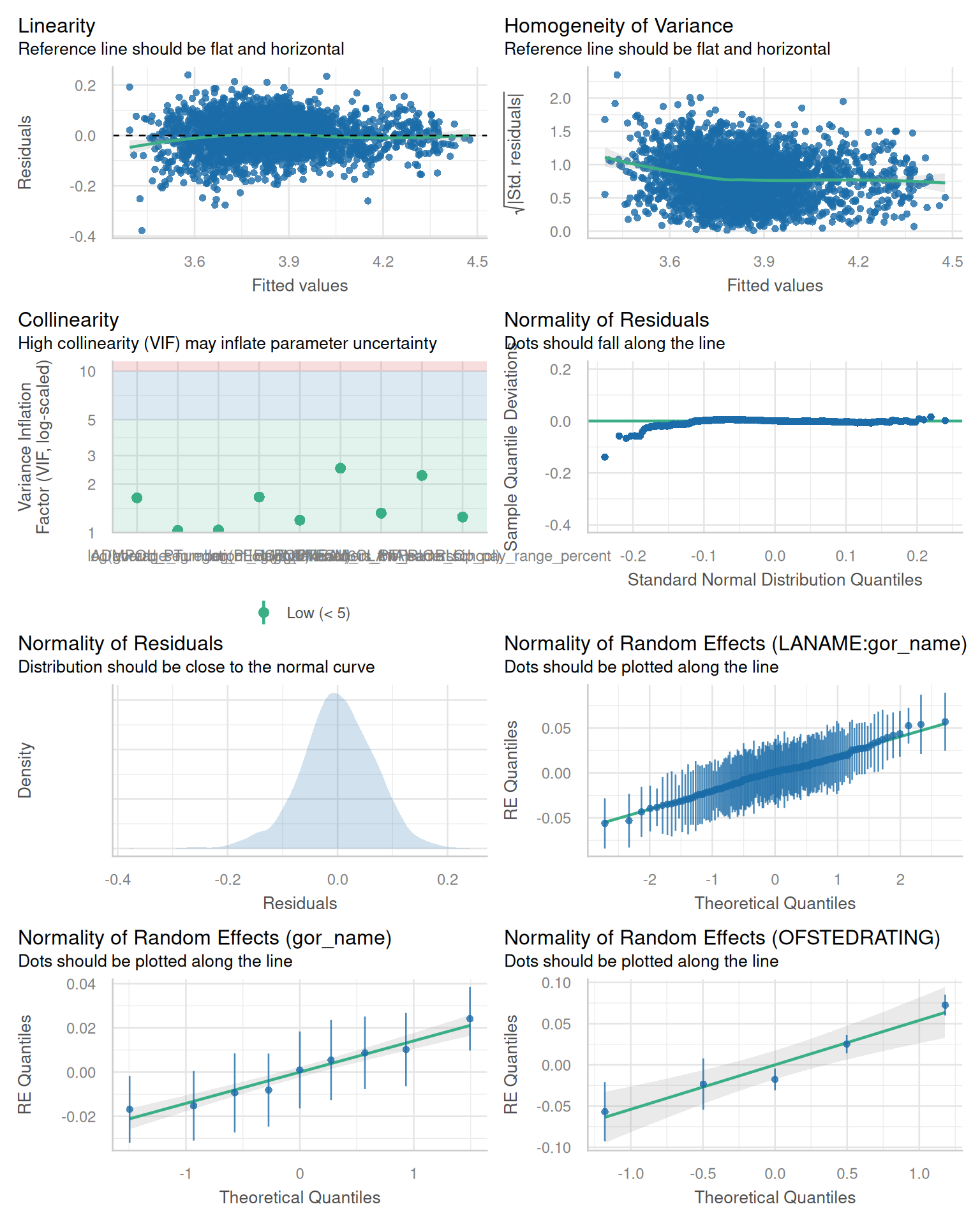
- We can also run some diagnostic plots to double check our model
adheres to the various assumptions required
```{r}
#| fig-width: 8
#| fig-height: 10
library(performance)
check_model(lme11)
```
### Interpreting your outputs
#### Random Effects
Turning the **Std.Dev** to an **approximate percentage difference**:
$\%= 100 \times (exp(SD) - 1)$. This gives a feel for how much
Attainment 8 changes for a **+1 SD higher** group intercept.
##### lme11 (Overall Attainment 8)
- **OFSTEDRATING**: Var = 0.0026, **SD = 0.051** → \~**+5.2%** per +1
SD
- **Region (gor_name)**: Var = 0.00024, **SD = 0.015** → \~**+1.5%**
- **LA within Region**: Var = 0.00066, **SD = 0.026** → \~**+2.6%**
- **Residual (school-level/unexplained)**: Var = 0.00471, **SD =
0.069** → \~**+7.1%**
Taking the sum of all the variances and dividing each individual
variance by this total gives the proportion of variance explained by
each random effect:
- **OFSTEDRATING**: **31.8%**
- **Region**: **2.9%**
- **LA within Region**: **8.1%**
- **Residual**: **57.3%**
- **Total variance**: 0.00822
While Ofsted Rating helps explain a lot of the variance - some 32%, most
of the variability is between individual schools within each group.
#### Fixed Effects
From **lme11 (overall attainment)**:
- **Most influential negative factors**:
- **Overall absence (log(PERCTOT))**: t ≈ -28.6 → Higher absence
strongly reduces attainment.
- **Prior low attainment (PTPRIORLO)**: t ≈ -28.5 → Schools with
more students entering low-performing do worse.
- **Proportion disadvantaged (log(PTFSM6CLA1A))**: t ≈ -17.1 →
Higher concentration of disadvantaged pupils lowers overall
attainment, but is much less important than the first two
factors.
- **Positive factors**:
- **Selective admissions (ADMPOL_PTSEL)**: t ≈ 9.0 → Selective
schools boost attainment.
- **Proportion EAL (log(PNUMEAL))**: t ≈ 7.5 → Linguistic
diversity slightly improves outcomes.
- **Teacher Stability (log(remained_in_the_same_school))**: t ≈
6.4 → Teachers remaining in the same school helps.
- **Moderate negatives**:
- **Teacher Absence days**: t ≈ -5.6.
- **Proportion of Teachers on Leadership pay %**: t ≈ -3.0.
- **Not significant**: Other admission policy, segregation index.
#### Overall Fit
Comparing the Observed and Fitted Values and calculating an $R^2$ value
from that, we can explain almost 90% of the variation in Attainment 8.
Schools in Brighton are mainly showing positive residuals - in other
words, performing much better than other similar schools in England with
similar profiles.
#### Benefits of LME over OLS
- **Accounts for clustering** (schools within OFSTED/regions).
<!-- -->
- **Shrinkage / "Partial pooling"** reduces overfitting and improves
predictions.
<!-- -->
- **Decomposes variance** across levels so we can see how, for
example, having a better Ofsted score might compare with lowering
absence.
<!-- -->
- **School-specific predictions** are more useful.
### Policy Interpretations and Using AI to help with the specific, contextual translation of outputs
- Below I have pulled out some of the schools in Brighton.
- One of the nice things about AI is it is very good at quickly
turning specific situations into policy-relevant examples using the
models we have just built. Have a look at the table below and I will
give you an example of a question that produces some very
interesting results:
```{r}
# Packages
library(dplyr)
library(tidyr)
library(stringr)
library(lme4)
library(tibble)
# --- Schools of interest ---
target_urns <- c("114581", "14608", "136164", "114580")
target_names <- c("Longhill High School", "Patcham High School", "Brighton Aldridge Community Academy", "Dorothy Stringer School")
# --- Variables of interest (from your final model) ---
vars_of_interest <- c(
"PERCTOT", # % overall absence
"PTFSM6CLA1A", # % disadvantaged
"PNUMEAL", # cohort size proxy
"PTPRIORLO", # prior low attainment
"ADMPOL_PT", # admissions policy
"gorard_segregation", # LA-level segregation
"remained_in_the_same_school", # total teachers remained_in_the_same_school
"teachers_on_leadership_pay_range_percent", # leadership pay %
"average_number_of_days_taken.x" # average sick days
)
# 1) Subset to target schools
schools_sub <- england_filtered_clean %>%
filter(URN %in% target_urns | SCHNAME.x %in% target_names) %>%
select(URN, SCHNAME.x, OFSTEDRATING, gor_name, LANAME, all_of(vars_of_interest)) %>%
distinct()
# 2) Extract random effects from lme11
re_list <- lme4::ranef(lme11)
# 2a) OFSTEDRATING random intercepts
re_ofsted <- re_list$OFSTEDRATING %>%
as.data.frame() %>%
rownames_to_column("OFSTEDRATING") %>%
rename(re_OFSTEDRATING = `(Intercept)`)
# 2b) gor_name random intercepts
re_gor <- re_list$gor_name %>%
as.data.frame() %>%
rownames_to_column("gor_name") %>%
rename(re_gor_name = `(Intercept)`)
# 2c) Nested random intercepts for LANAME:gor_name
grp_names <- names(re_list)
lan_gor_grp <- grp_names[grepl("LANAME.*gor_name|gor_name.*LANAME", grp_names)][1]
if (grepl("^LANAME", lan_gor_grp)) {
re_lan_gor <- re_list[[lan_gor_grp]] %>%
as.data.frame() %>%
rownames_to_column("key") %>%
separate(key, into = c("LANAME", "gor_name"), sep = ":", remove = TRUE) %>%
rename(re_LANAME_gor = `(Intercept)`)
} else {
re_lan_gor <- re_list[[lan_gor_grp]] %>%
as.data.frame() %>%
rownames_to_column("key") %>%
separate(key, into = c("gor_name", "LANAME"), sep = ":", remove = TRUE) %>%
rename(re_LANAME_gor = `(Intercept)`) %>%
relocate(LANAME, gor_name)
}
# 3) Join random effects and compute total RE
schools_with_re <- schools_sub %>%
left_join(re_ofsted, by = "OFSTEDRATING") %>%
left_join(re_gor, by = "gor_name") %>%
left_join(re_lan_gor, by = c("LANAME", "gor_name")) %>%
mutate(RE_total = re_OFSTEDRATING + re_gor_name + re_LANAME_gor)
# 4) Predictions (log scale -> back-transform)
pred_log_fixed <- predict(lme11, newdata = schools_with_re, re.form = NA)
pred_log_cond <- predict(lme11, newdata = schools_with_re, re.form = NULL)
# Bias correction
sigma2 <- sigma(lme11)^2
bias_factor <- exp(0.5 * sigma2)
schools_with_re <- schools_with_re %>%
mutate(
pred_fixed = exp(pred_log_fixed),
pred_cond = exp(pred_log_cond),
pred_cond_bc = pred_cond * bias_factor
)
# 5) Display table
row_order <- c(
"PERCTOT", "PTFSM6CLA1A", "PNUMEAL", "PTPRIORLO", "ADMPOL_PT",
"gorard_segregation", "remained_in_the_same_school", "teachers_on_leadership_pay_range_percent", "average_number_of_days_taken.x",
"RE: OFSTEDRATING", "RE: gor_name", "RE: LANAME:gor_name", "RE: total",
"Baseline ATT8 (fixed only)", "Baseline ATT8 (incl. RE)", "Baseline ATT8 (incl. RE, bias-corrected)"
)
display_tbl <- schools_with_re %>%
transmute(
SCHNAME.x,
PERCTOT = formatC(PERCTOT, format = "f", digits = 1),
PTFSM6CLA1A = formatC(PTFSM6CLA1A, format = "f", digits = 1),
PNUMEAL = formatC(PNUMEAL, format = "f", digits = 1),
PTPRIORLO = formatC(PTPRIORLO, format = "f", digits = 1),
ADMPOL_PT = as.character(ADMPOL_PT),
gorard_segregation = formatC(gorard_segregation, format = "f", digits = 3),
remained_in_the_same_school = formatC(remained_in_the_same_school, format = "f", digits = 3),
teachers_on_leadership_pay_range_percent = formatC(teachers_on_leadership_pay_range_percent, format = "f", digits = 1),
average_number_of_days_taken.x = formatC(average_number_of_days_taken.x, format = "f", digits = 3),
`RE: OFSTEDRATING` = formatC(re_OFSTEDRATING, format = "f", digits = 3),
`RE: gor_name` = formatC(re_gor_name, format = "f", digits = 3),
`RE: LANAME:gor_name` = formatC(re_LANAME_gor, format = "f", digits = 3),
`RE: total` = formatC(RE_total, format = "f", digits = 3),
`Baseline ATT8 (fixed only)` = formatC(pred_fixed, format = "f", digits = 1),
`Baseline ATT8 (incl. RE)` = formatC(pred_cond, format = "f", digits = 1),
`Baseline ATT8 (incl. RE, bias-corrected)` = formatC(pred_cond_bc, format = "f", digits = 1)
) %>%
pivot_longer(cols = -SCHNAME.x, names_to = "Variable", values_to = "Value") %>%
pivot_wider(names_from = SCHNAME.x, values_from = Value) %>%
mutate(Variable = factor(Variable, levels = row_order)) %>%
arrange(Variable)
knitr::kable(
display_tbl %>% arrange(Variable),
align = "l"
)
```
::: {.callout-tip title="Question for my AI"}
Given the outputs from the model above and the specific detail the
variables for each school. Considering Longhill and Patcham schools, can
you show what percentage point change would be required for each
variable to affect an increase of 5 more Attainment 8 points?
:::
| Variable | Change (unit) | Longhill Current | Longhill Required Δ | Patcham Current | Patcham Required Δ |
|------------|------------|------------|------------|------------|------------|
| **PERCTOT** (overall absence) *(log)* | pp | 13.4 | **−5.9 pp** | 9.2 | **−3.3 pp** |
| **PTFSM6CLA1A** (% disadvantaged) *(log)* | pp | 29.0 | −24.3 pp | 19.0 | −14.2 pp |
This output is highly illustrative and shows that given the different
starting points for each school, not only are different levels of effort
required to reach the same outcomes (Patcham would need to reduce
absence by fewer percentage points to achieve the same 5 point
attainment 8 increase), but we can see how much more effective reducing
absence is than reducing levels of disadvantage - with Longhill needing
to reduce its levels of disadvantaged students down from 29% to to 5% to
achieve the same point score improvement.
Given the policy levers that Brighton and Hove Council tried to pull,
this is very important information.
### Task
See if you can experiment with feeding your own tame AI (Copilot/Chat
GPT, ClaudeAI, Gemini etc. will all do this slightly differently and
with greater or lesser success) with your model outputs and some specifics about some schools or local authorities, and then ask some questions of it to aid in some insightful interpretation.
::: {.callout-caution}
I shouldn't need to tell you to proceed with caution when asking an AI to interpret your outputs. It will get things wrong so don't just copy and paste! Interrogate, ask follow-up questions. Ask for reinterpretations or to explain specific features.
:::
## Extension Activities
I think you've probably had enough at this point, but you will hopefully find many uses for these models in your future work.
```{r}
#| eval: false
#| include: false
lme12 <- lmer(
log(ATT8SCR_FSM6CLA1A) ~ log(PTFSM6CLA1A) + log(PERCTOT) + log(PNUMEAL) +
PTPRIORLO + ADMPOL_PT + gorard_segregation +
log(remained_in_the_same_school) + teachers_on_leadership_pay_range_percent + log(average_number_of_days_taken.x) +
(1 | OFSTEDRATING) + (1 | gor_name/LANAME),
data = england_filtered_clean,
na.action = na.exclude
)
summary(lme12)
summary(lme11)
```