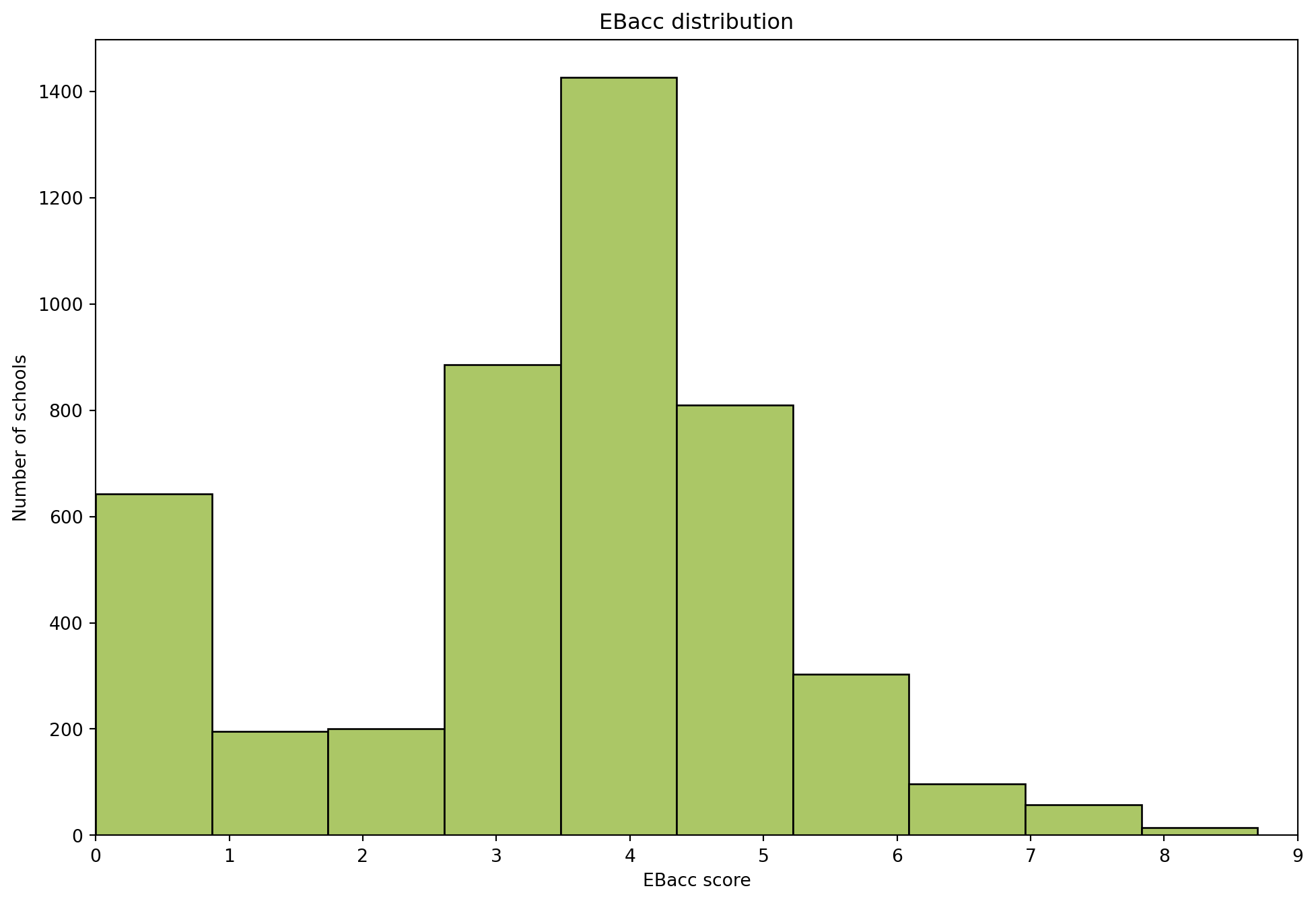
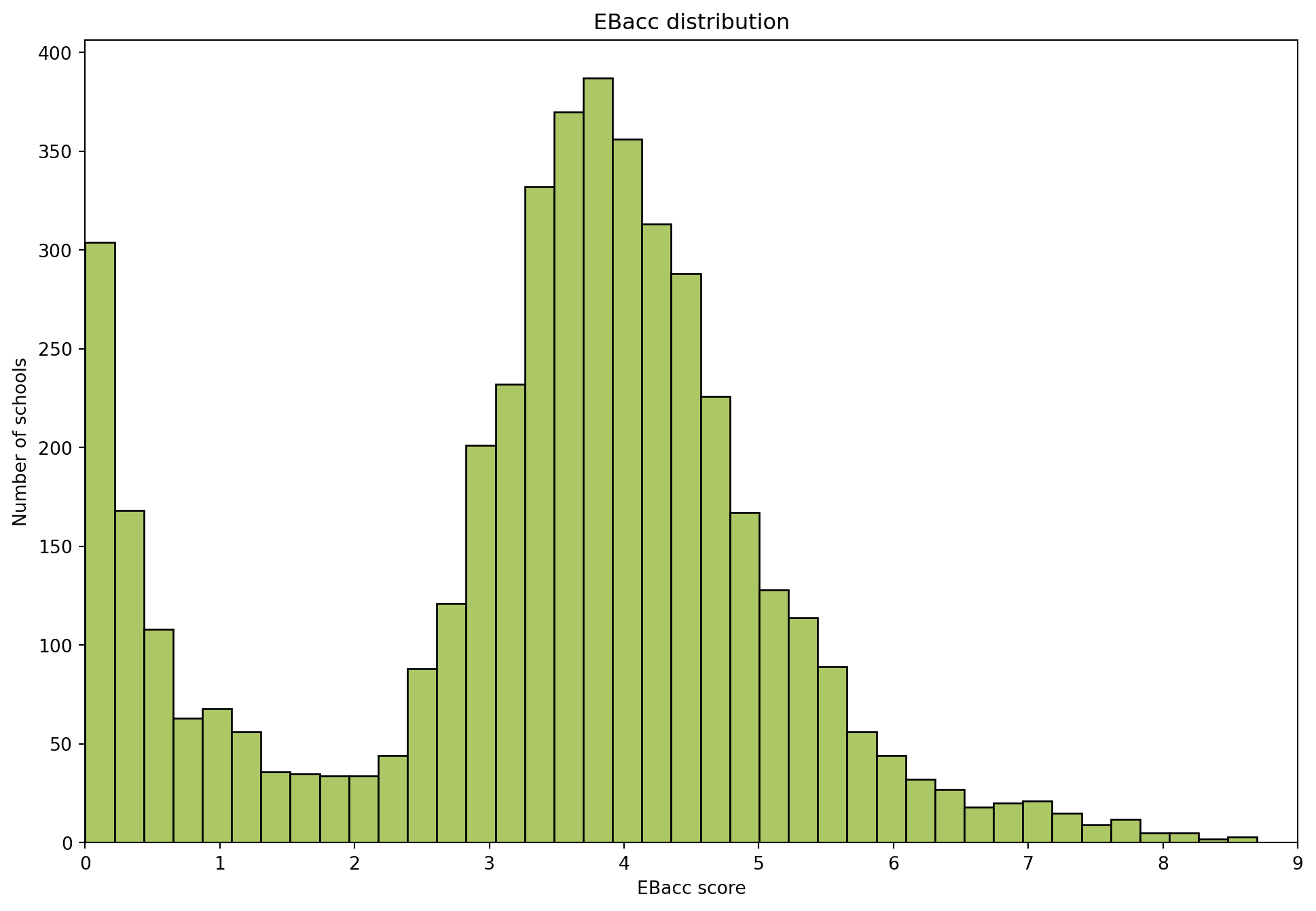
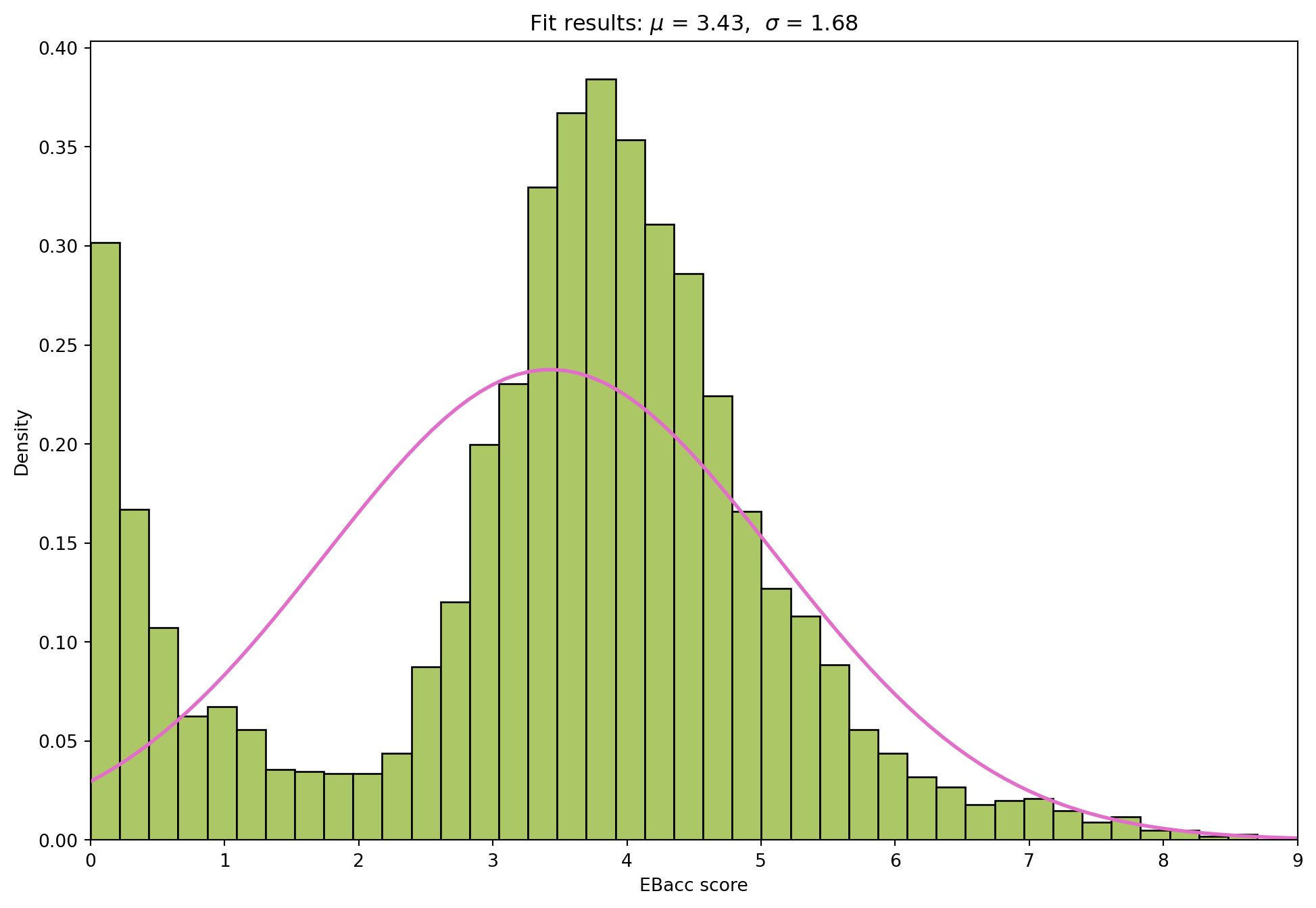
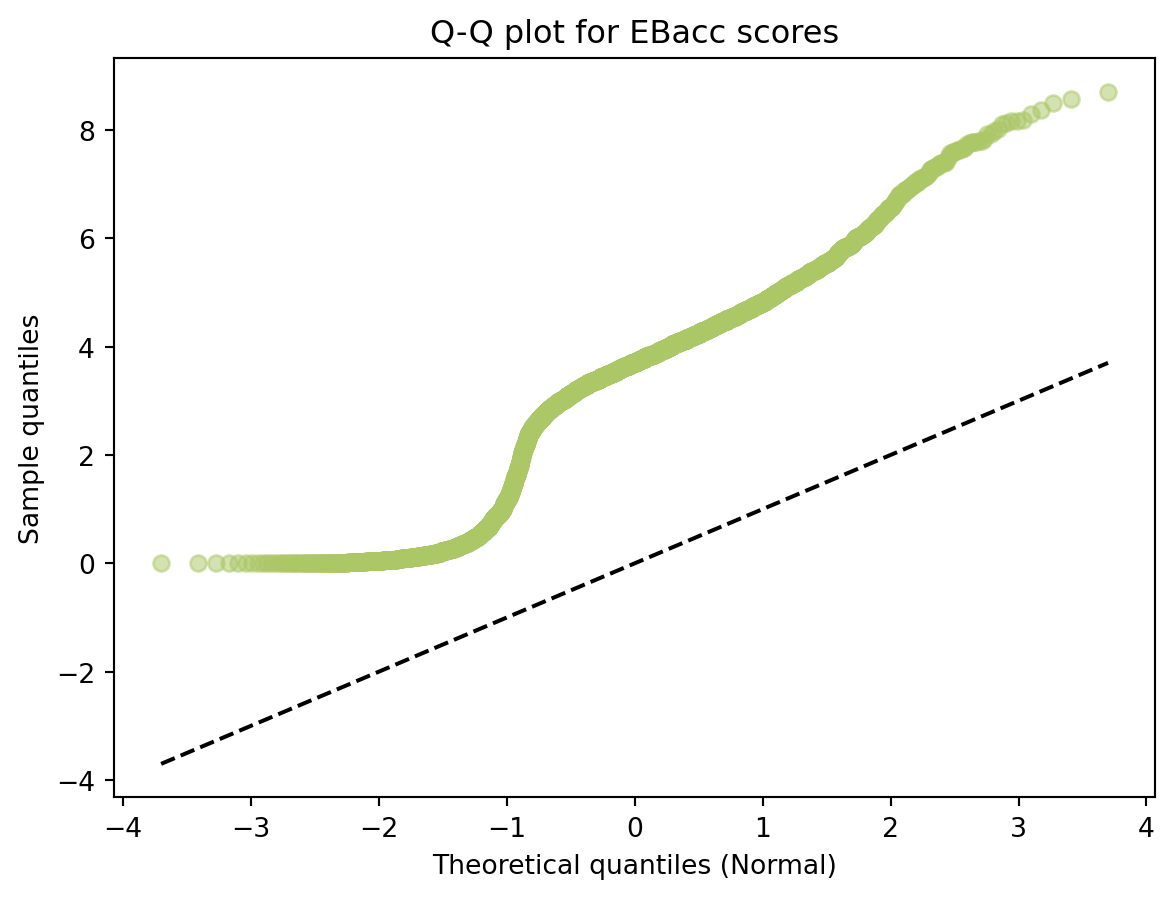
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
# Read CSV file, handling common missing value entries
na_vals = ["", "NA", "SUPP", "NP", "NE", "SP", "SN", "SUPPMAT"]
df_ks4 = pd.read_csv('https://raw.githubusercontent.com/huanfachen/QM/refs/heads/main/sessions/L2_data/england_ks4final.csv',
na_values = na_vals
)
info_cols = ['RECTYPE', 'LEA', 'SCHNAME', 'TOTPUPS']
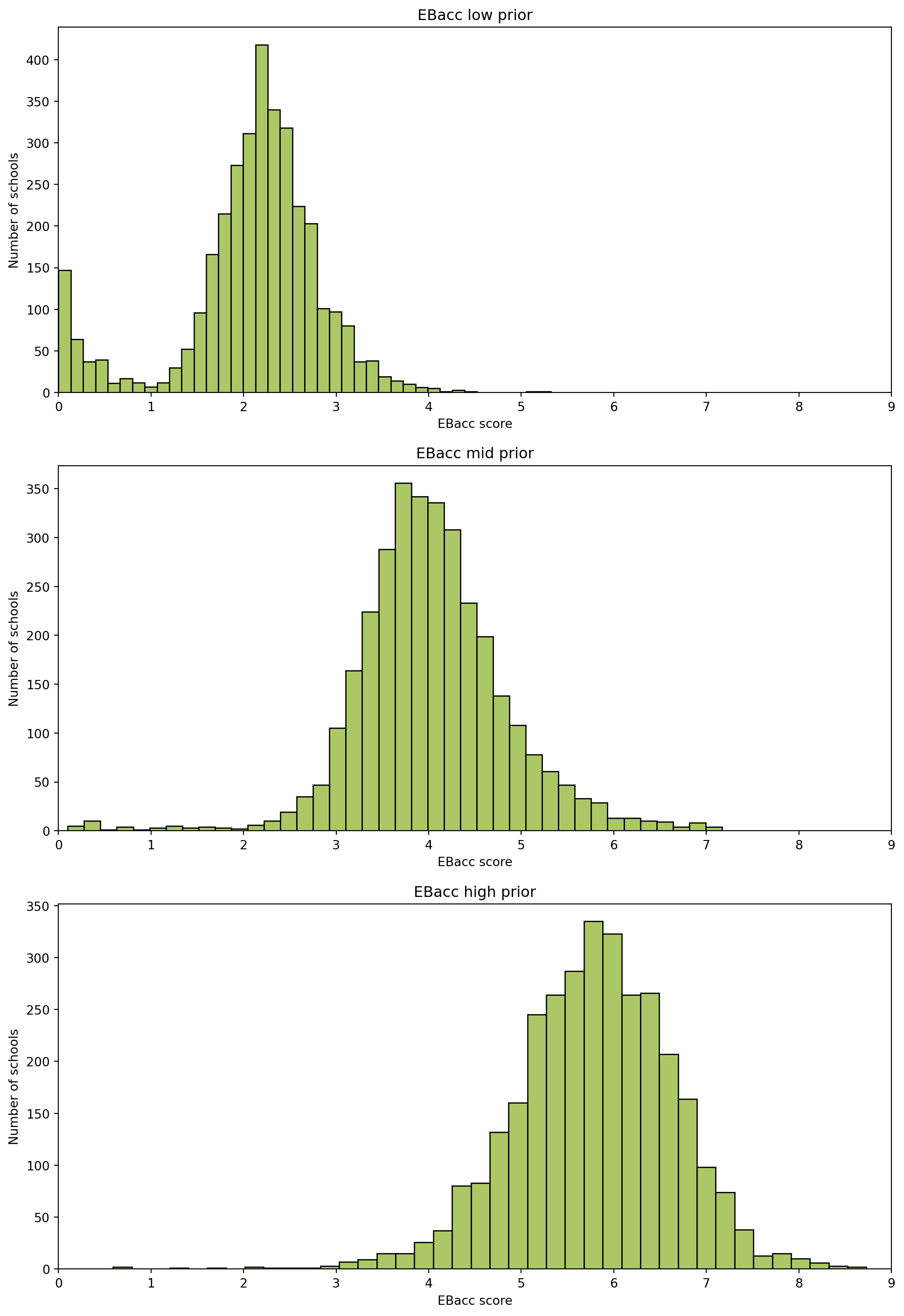
ebaccs_cols = ['EBACCAPS', 'EBACCAPS_LO', 'EBACCAPS_MID', 'EBACCAPS_HI']
df = df_ks4[info_cols + ebaccs_cols]
# work on a copy of the dataframe to avoid SettingWithCopyWarning
df = df.copy()
df.head()/tmp/ipykernel_34779/1412533053.py:7: DtypeWarning:
Columns (75,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,144,145,146,147,148,149,150,151,152,177,178,179,180,181,182,183,186,187,188,189,190,191,192,194,195,196,198,199,200,202,203,204,206,207,208,210,211,212,214,215,216,218,219,220,222,223,224,230,233,234,235,236,237,238,239,242,243,244,245,246,247,248,251,252,253,254,255,256,257,266,267,268,269,270,271,272,281,282,283,284,285,286,287,296,297,298,299,300,301,302,311,312,313,314,315,316,317,335,336,337,340,341,342,345,346,347,366,367,368,369,370,371,372,373,374,375,376,377,378,379,380,381,382,383,384,385,386,387,388,389,390,391,392,393,394,395,396,397,398,399,400,401,402,403,404,405,406,407,408,409,410) have mixed types. Specify dtype option on import or set low_memory=False.
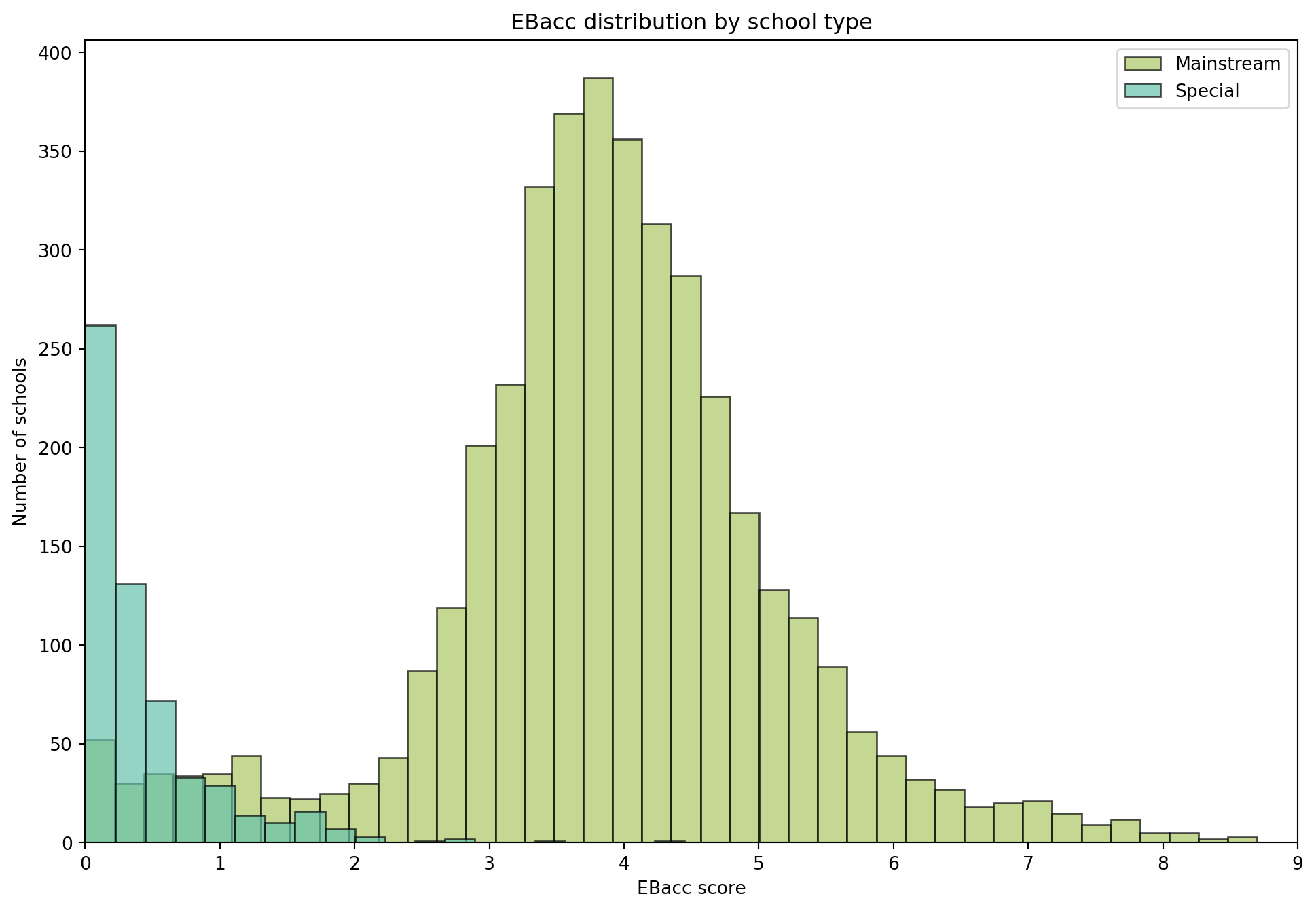
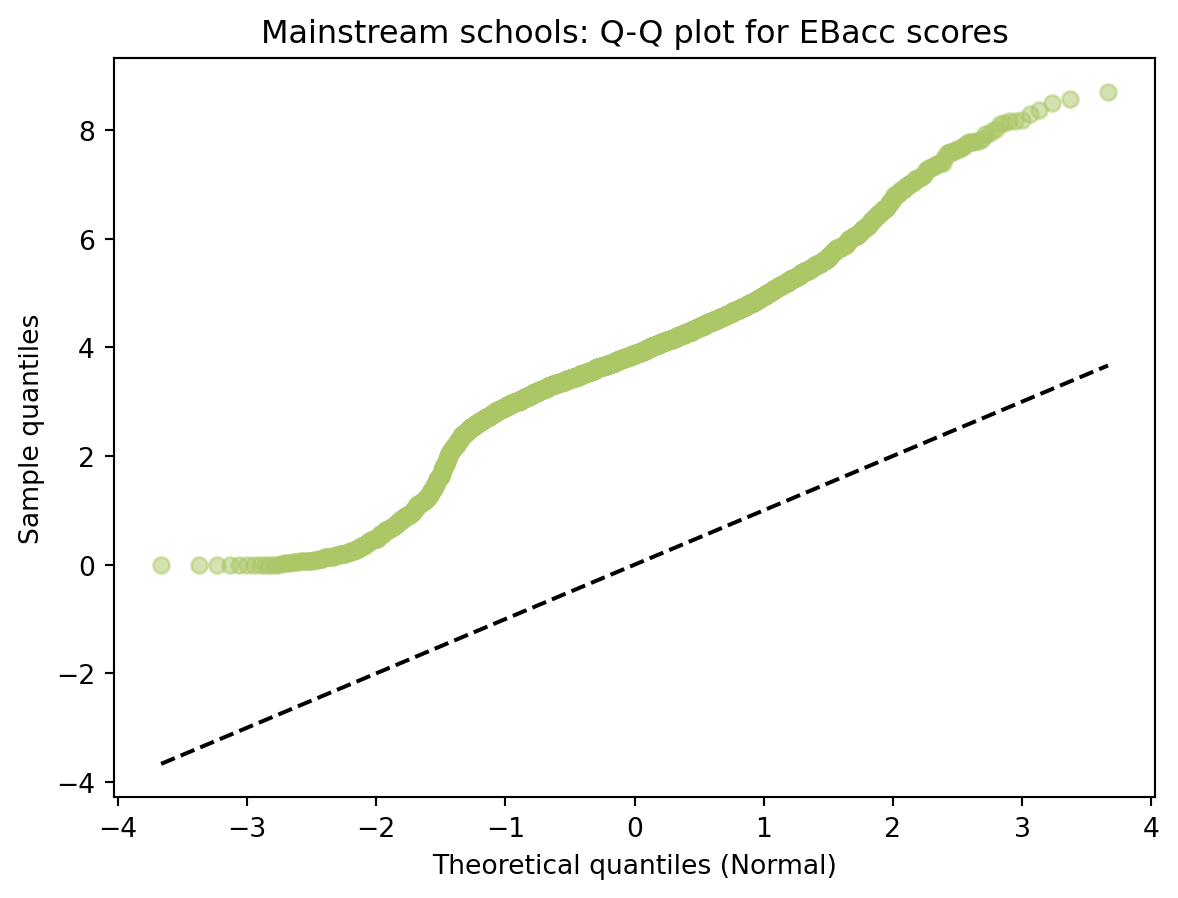
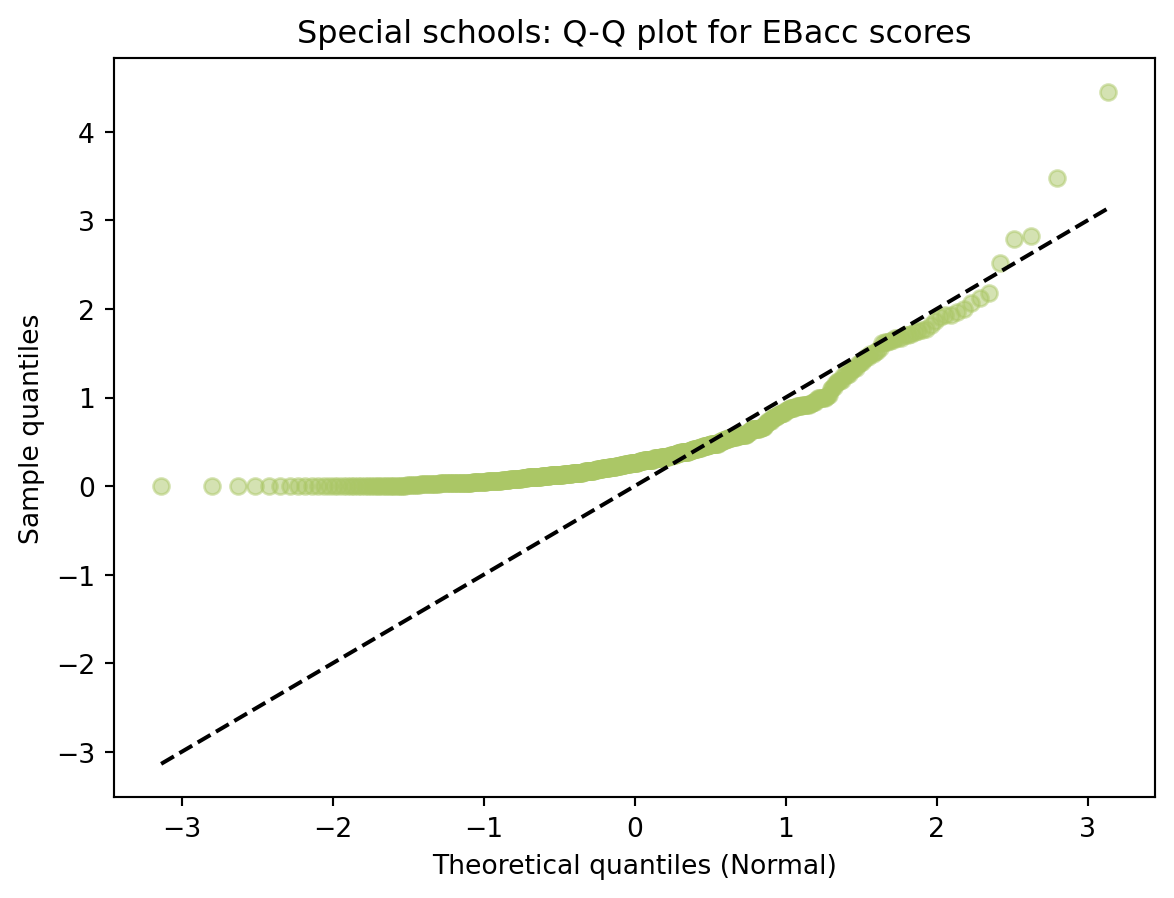
| RECTYPE | LEA | SCHNAME | TOTPUPS | EBACCAPS | EBACCAPS_LO | EBACCAPS_MID | EBACCAPS_HI | |
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 201.0 | City of London School | 1045 | 2.10 | NaN | NaN | NaN |
| 1 | 1 | 201.0 | City of London School for Girls | 739 | 1.51 | NaN | NaN | NaN |
| 2 | 1 | 201.0 | David Game College | 365 | 0.56 | NaN | NaN | NaN |
| 3 | 4 | 201.0 | NaN | 0 | NaN | NaN | NaN | NaN |
| 4 | 1 | 202.0 | Acland Burghley School | 1163 | 4.62 | 1.91 | 3.81 | 6.87 |